ABSTRACT
Protein tyrosine nitration is an important post-translational modification. A variety of nitrated proteins are reported in Arabidopsis leaves and seedlings, sunflower hypocotyls, and pea roots. The identities of nitrated proteins are species-/organ-specific, and chloroplast proteins are most nitratable in leaves. However, precise mechanism is unclear. Here, we investigated nitroproteome in tobacco leaves following exposure to nitrogen dioxide. Proteins were extracted, electrophoresed and immunoblotted using an anti-3-nitrotyrosine antibody. Mass spectrometry and FASTA search identified for the first time an exclusive nitration of pathogenesis-related proteins, PR-1, PR-3 and PR-5, which are reportedly located in the apoplast or the vacuole. Furthermore, Tyr36 of thaumatin-like protein E2 was identfied as a nitration site. The underlying mechanism and physiological relevance are discussed.
KEYWORDS: Apoplast, defense/immunity-related proteins, nitrogen dioxide, pathogenesis-related proteins, PR-1, PR-3, PR-5, protein tyrosine nitration, tobacco, vacuole
Protein tyrosine nitration is a covalent post-translational protein modification that plays a critical role in cell physiological processes, including cellular signaling.1-5 Plant proteomic studies have identified a variety of nitrated proteins in the leaves and seedlings of Arabidopsis [Arabidopsis thaliana (L.) Heynh.],6-9 the hypocotyls of sunflower (Helianthus annuus L.),10,11 and the roots of pea (Pisum sativum L.).12 Each of these organs has shown nitration of a distinctly different set of proteins, and thus nitration is considered specific for organs analyzed.
Arabidopsis leaves following exposure to peroxynitrite,6 high light7 or nitrogen dioxide (NO2)9 have identified nitration of 8, 14 and 6 different proteins, respectively. Interestingly, all these proteins but one cytosol protein6 are located in the chloroplast. Exposure to NO2 induces nitration of PsbO and PsbP, extrinsic proteins of photosystem II (PSII), and 4 non-PSII proteins, including peroxiredoxin II E (PRX IIE).9 Furthermore, the proteomic analysis of Arabidopsis seedlings grown under unstressed condition also showed nitration of PsbP.8 Thus, chloroplasts may be a major target organelle of protein nitration in leaves. However, the precise regulatory mechanism controlling protein nitration remains unelucidated.
In order to gain insight into this aspect of protein nitration, we therefore performed proteomic identification of nitrated proteins in the leaves of tobacco, a fourth species, following exposure to NO2. We found for the first time that nitration was exclusive to pathogenesis-related (PR)-1 (PR-1A and PR-1B), PR-3 (chitinases PR-P and PR-Q) and PR-5 (thaumatin-like proteins E22 and E2) proteins. All of these proteins are reportedly localized in the apoplast or vacuole. Furthermore, Tyr36 of thaumatin-like protein E2 was identfied as a nitration site as shown in the followings.
Tobacco plants (Nicotiana tabacum cv. Xanthi XHFD8) were grown for 9 weeks and used in this study. They were exposed to NO2 (4.0 ± 0.1 ppm) in an NO2 chamber for 8 h in light (100 µmol m−2 s−1) and from them protein was extracted as described in Supplemental materials and methods.
One-dimensional polyacrylamide gel electrophoresis (1D PAGE) was carried out according to Görg et al. (1985).13 Immunoblotting was performed using a rabbit polyclonal anti-NT antibody (Upstate Biotechnology, Lake Placid, NY, USA), and a goat anti-rabbit IgG secondary antibody conjugated with horseradish peroxidase(VECTOR Lab., Burlingame, CA, USA) as described in Supplemental materials and methods. Proteins extracted from tobacco leaves exposed or not exposed to NO2 showed several NT-immunopositive bands or very faint bands, respectively, in 1D immunoblot gels (Fig. S1). Immunoblot analysis using nitrated bovine serum albumin (BSA) as a control confirmed the specificity of the anti-NT antibody (Fig. S1).
Two dimensional PAGE (2D PAGE) was carried out according to Görg et al. (1985)13 with some modifications as described elsewhere.14 For iodoacetamide treatment, after isoelectric focusing (IEF), the immobilized pH gradient (IPG) strips were equilibrated for 30 min in the dark with SDS buffer containing 2% (w/v) SDS, 50 mM Tris-HCl (pH6.8), 6 mM urea, 30% (w/v) glycerol, 25 mg/mL iodoacetamide and a trace of bromophenol blue, and applied to SDS gels for resolution in 2D. The 2D PAGE gels were stained with SYPRO Ruby stain (Bio-Rad), and protein spots were visualized using a fluorescence scanner (Bio-Rad Molecular Imager FX) and the PDQuest software (Bio-Rad), as described in Supplemental information. Immunoblotting was performed using the anti-NT antibodies, as described in Supplemental materials and methods.
Approximately 600 protein spots were detected on a SYPRO Ruby-stained 2D gel (Fig. 1B) of proteins extracted from exposed tobacco leaves. Among them, 8 spots (spots 1–8) were detected by an anti-NT antibody (Fig. 1A). These proteins were in the pI range of 4.7–6.2 and the molecular mass range of 18–37 kDa. Six spots (1–4, 7, and 8) were further investigated, while spots 5 and 6 showed no detectable peptides by mass spectrometry.
Figure 1.
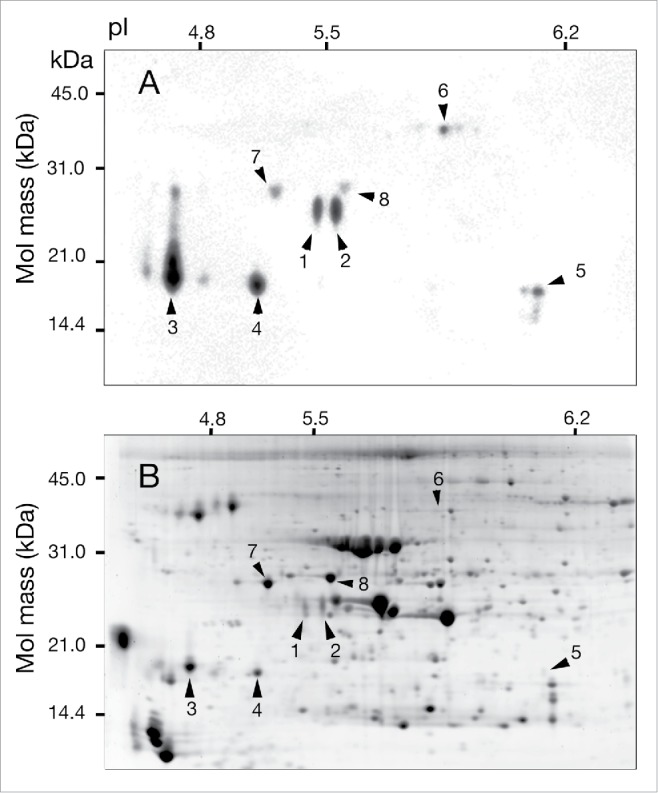
Two-dimensional PAGE of proteins from tobacco leaves exposed to NO2. Nine-week-old tobacco plants (Nicotiana tabacum cv Xanthi XHFD8) were exposed to NO2 at 4.0 ± 0.4 ppm for 8 h in light. Proteins were extracted from the exposed plants and subjected to 2D PAGE. Protein spots were visualized by staining with a polyclonal anti-3-NT antibody (A) and SYPRO Ruby (B). Nine-hundred micrograms of protein were loaded on each gel.
These spots were individually excised and in-gel digested by trypsin (Promega) according to Shevschenko et al. (1996).15 The tryptic digests were analyzed using a hybrid quadrupole time-of-flight mass spectrometer (Q-TOF MS) (Micromass, Manchester, UK) in the MS and MS/MS mode according to Kristensen et al. (2000).16 The Q-TOF mass spectra (parent ions) of the tryptic peptides were first obtained in the MS mode. The parent ions of interest were then subjected to sequence analysis in the MS/MS mode to determine amino acid sequences (designated internal sequences). The MS and MS/MS data were subjected to a FASTA search against the SwissProt and GenBank databases using the following parameters: one missed cleavage was allowed; oxidation of methionine (+15.99 Da) and pyroglutamination of the amino-terminal glutamine (−17.06 Da) were set as variable modifications, and carbamidomethylation of cysteine (+57.02 Da) was set as a fixed modification. Additionally, some peptides were analyzed using a matrix- assisted laser desorption ionization-time of flight mass spectrometer (MALDI-TOF MS) according to Matsuo et al. (2006)17 as described in Supplemental materials and methods. In this study, unless noted otherwise, all MS analyses were performed using Q-TOF MS. The results are summarized in Table 1. Detailed analyses of the data obtained by MS analyses are available in Supplemental results.
Table 1.
Nitrated proteins identified by Q-TOF MS analysis of tryptic peptides of proteins extracted from tobacco leaves following exposure to NO2.
| Spot no. | Charge1)status | m/z1)measured | m/z1)calculated | Sequence2) | Identity | Protein name | pI (Mw3)) measured | pI (Mw3)) calculated | UniProtKB accession number |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 1015.51 | 1015.01 | 50LDSGQSWSINVNPGTVQAR68 | 19/19 | E22 | 5.6 (25) | 4.94 (21.7) | P13046 |
| 2 | 2 | 1079.61 | 1078.55 | 49QLNSGQSWSINVNPGTVQAR68 | 20/20 | E2 | 5.7 (25) | 4.87 (21.6) | P07052 |
| 3 | 2 | 871.89 | 872.40 | 31QNSQQDYLDAHNTAR45 | 15/15 | PR-1A, B, C | P08299, P07053, P09042 | ||
| 2 | 1483.42 | 1483.60 | 143VQCNNGGYVVSCNYDPPGNYRGESPY168 | 26/26 | PR-1A | 4.7 (19) | 4.57 (15.2) | P08299 | |
| 4 | 2 | 871.93 | 872.40 | 31QNSQQDYLDAHNTAR45 | 15/15 | PR-1A, B, C | P08299, P07053, P09042 | ||
| 2 | 1429. | 1429.65 | 143VKCNNGGYVVSCNYDPPGNVIGQSPY168 | 26/26 | PR-1B | 5.3 (18) | 4.89 (15.1) | P07053 | |
| 7 | 2 | 1065.99 | 1066.53 | 25QGIGSIVTNDLFNEMLKNR43 | 19/19 | PR-P | 5.4 (27) | 5.00 (24.8) | P17513 |
| 2 | 1073.94 | 1075.59 | 25QGIGSIVTNDLFNEMLKNR43 | 19/19 | PR-P | P17513 | |||
| 2 | 1137.90 | 1138.47 | 230YCGMLNVAPGDNLDCYNQR248 | 19/19 | PR-P | P17513 | |||
| 8 | 2 | 714.35 | 714.82 | 65SFPGFGTTGDDTAR78 | 14/14 | PR-Q | 5.7 (27) | 5.22 (25.0) | P17514 |
Shown are charge status and the measured and calculated m/z values of parent ions detected in the Q-TOF mass spectra of trypsin digests of NT-positive protein spots from tobacco leaves following exposure to NO2.
Shown is the amino acid sequence of the tryptic fragment. Internal sequences determined by the Q-TOF MS analysis are shown in bold letters. Q; Pyroglutamine, C; carbamidomethylated cysteine, M; oxidized methionine. Sequences were subjected to a FASTA search against the Swiss-Prot database to identify proteins.
Mw values are shown in kDa.
The MS mode analysis of the trypsin digest of spot 1 detected a doubly charged [M + 2H]2+parent ion at m/z 1015.51, and the subsequent MS/MS mode analysis determined an internal sequence consisting of 17 residues (Table 1). A detailed analysis of these data (see Supplemental results) identified spot 1 as thaumatin-like protein E22 (Table 1).
A doubly charged parent ion at m/z 1079.61 was detected in the MS spectra of the trypsin digests of spot 2, and from this parent ion, a 17-residue internal sequence identical to that of spot 1 was identified (Table 1). A detailed analysis of these data (see Supplemental results) identified spot 2 was as thaumatin-like protein E2 (Table 1).
E22 and E2 are isoforms of the group 5 PR proteins in tobacco.18,19 Both the PR-5 proteins share significant amino acid sequence homology with the sweet-tasting protein, thaumatin, in the fruits of the tropical plant Thaumatococcus daniellii and are thus referred to as thaumatin-like proteins.20
In the MS spectra of the trypsin digests of spot 3, 2 doubly charged parent ions at m/z 871.89 and 1483.42 were detected, from which 13- and 7-residue internal sequences, respectively, were determined (Table 1). A detailed analysis of these data (see Supplemental results) identified spot 3 as PR-1A.
In the MS spectra of the trypsin digests of spot 4, 2 doubly charged parent ions at m/z 871.93 and 1429.37 were detected, which 13- and 12-residue internal sequences, respectively, were determined (Table 1). A detailed analysis of these data (see Supplemental results) identified spot 4 as PR-1B. PR-1A and PR-1B are localized in the apoplast (extracellular space) and/or vacuole, depending on the cell type, in tobacco.21,22 PR-1 proteins are the most abundant PR protein.18,19
In the MS spectra of the trypsin digests of spot 7, 3 doubly charged parent ions were detected at m/z 1065.99, 1073.94 and 1137.90 (Table 1). The internal sequences determined from the former 2 ions consisted of 16 identical residues. A detailed analysis of these data together with peptide finger printing (PMF) analysis using MALDI-TOF MS (see Supplemental results) identified spot 7 as PR-P protein.
In the MS spectra of tryptic digests of spot 8, a parent ion at m/z 714.35 was detected, from which a 12-residue internal sequence was determined. A detailed analysis of these data together with PMF analysis (see Supplemental results) identified spot 8 as PR-Q protein (Table 1). PR-P and PR-Q are acidic chitinases of the group 3 PR proteins in tobacco (Table 1).18,19 Both PR-3 proteins reportedly display antifungal activities and are located in the apoplast.18
A basic endochitinase and a Populus trichocarpa chitinase family protein are nitrated in pea roots.12 Although these chitinase species are distinct from tobacco acidic PR-3 chitinases (PR-P and PR-Q) (see above), it is noteworthy that chitinase species are a common target for nitration in various organs of different species.
The PR proteins identified in this study have 5 to 11 tyrosine residues per molecule (UniProt). However, no nitrated peptides were detected in the tryptic digests of tobacco leaf proteins by the present Q-TOF analyses. We therefore analyzed the nitrated peptides using MALDI-TOF MS according to Matsuo et al. (2006).17 Tobacco leaves were exposed to 4.0 ± 0.4 ppm 15N-labeled NO2(51.6 atom% 15N) for 8 h in light.14 Leaf proteins were extracted from exposed plants and subjected to 2D PAGE and immunoblotting using the anti-NT antibody as described above, except that iodoacetamide was omitted from the SDS buffer so that cysteine residues were not carbamidomethylated. Formation of a cysteine acrylamide adduct (+71.08 Da) was set as a variable modification. Since nitration results in a mass shift of +45 or +46 Da (replacement of H with a NO2 or 15NO2 group, respectively), we searched pairs of ions having such a mass shift in the spectra of the in-gel digests of spot 2 digested by trypsin and V8 protease (Sigma).
A pair of 2 singly charged ions at m/z 2023.3 and 2069.3 was found (Fig. 2). The Mascot database search indicated that these ions correspond to residues 30–48 (30IVNQCTYTVWAAASPGGGR48) of E2, without or with one nitrated tyrosine, respectively. Formation of an acrylamide adduct at Cys34 was postulated in both ions. The nitration or replacement of H with a 15NO2 group resulted in a mass shift of +46 Da from m/z 2023.3 to 2069.3 (Fig. 2), identifying Tyr36 of E2 as the site of nitration. However, all attempts to determine nitration site identification in nitrated proteins other than E2 have been in vain. Reason(s) for this is unclear.
Figure 2.

Detection of 3-NT-containing peptides of thaumatin-like protein E2 by MALDI-TOF MS. Proteins were extracted from tobacco leaves exposed to 4.0 ± 0.4 ppm 15NO2(51.6 atom% 15N), and subjected to 2D PAGE followed by SYPRO RUBY staining and immunoblotting using an anti-NT antibody. Spot 2 on the SYPRO-RUBY-stained gel corresponding to the signal obtained from immunoblot analysis was excised, in-gel digested with trypsin and V8 protease, and identified by MALDI-TOF/TOF MS. A Mascot search indicated that 2 singly charged [M + H]+ ion peaks at m/z 2023.3 and 2069.3 correspond to residues 30–48 (30IVNQCTYTVWAAASPGGGR48) of E2, without or with one nitrated tyrosine, respectively. Nitration (replacement of H with an 15NO2 group) resulted in a mass shift of +46 Da.
It is possible that exogenously applied NO2 enters cells and nitrate proteins. It is also possible that exogenously applied NO2 elevate nitrosative stress in cells to stimulate production24 of endogenous NO2, which nitrate proteins. Thus, whether the nitro groups of the nitrated proteins following NO2 exposure are derived from the exogenously supplied NO2 or from endogenous24 NO2 has so far been unclear. Our present finding of a mass shift of +46 Da, in turn, indicates that the nitro group in nitrated proteins produced by NO2-induced nitration treatment is derived from exogenously supplied NO2.
Our present study showed that of ∼600 protein spots 6 NT-positive proteins were all identified as PR proteins, PR-1A, PR-1B, PR-P, PR-Q, thaumatin-like protein E2 and thaumatin-like protein E22. Protein tyrosine nitration is not a random chemical process, but is instead a selective process in which only specific proteins are nitrated,1,2 and neither the abundance of a protein nor the abundance of tyrosine residues in a given protein can predict whether it is a target for protein tyrosine nitration.1,2 However, this exclusive nitration of PR proteins in tobacco leaves was a totally unexpected observation. Nonetheless, we thought our present finding important in light of the reported fact that PR proteins play a vital role in plant defense/immune systems.17,18 However, in order to clarify whether apoplastic / vacuolar proteins are nitratale, immuno-electron microscopic evidence25 to show localization of nitrated proteins in the apoplast / the vacuole in tobacco leaves following exposure to NO2 is absolutely required.
PR proteins are defined as inducible defense-related proteins.17,18 They are inducible upon pathogen infection and biotic/abiotic stress and play a role in the surveillance mechanism in the plant immune system.18 They are currently classified into 17 families.17,18 Those PR proteins whose nitration was identified in the present study are inducible by abiotic stresses such as nitric oxide (NO),26 salicylic acid27 and ozone28 in tobacco leaves. They may also have been induced upon exposure to NO2.
It should be noted, however, that the concentration of NO2 used in the present study is ∼100-fold higher than those (10–200 ppb) used in NO2-mediated regulation of plant growth.9,29,30 Therefore, further investigations are required to establish the physiological relevance of the present observations on nitration of these PR proteins. Future studies will focus on site-directed mutagenesis of Tyr36 of E2, a nitration site identified in this study, followed by introduction of a mutated E2 gene in tobacco plants. The engineered and non-engineered control plants will be grown in the presence or absence of physiological concentrations of NO2 to explore the relationship between nitration of defense-related PR proteins and NO2-mediated growth regulation.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Financial support from the Nippon Life Insurance Foundation (to MT) is acknowledged. This research was supported in part by the Research for the Future Program, Japanese Society for the Promotion of Science (JSPS-RFTF96L00604) and by a Grant-in-Aid for Creative Scientific Research (no. 13GS0023) from the Japan Society for the Promotion of Science (to HM).
References
- 1.Ischiropoulos H. Protein tyrosine nitration—An update. Arch Biochem Biophys 2009; 484:117-21; PMID:19007743; http://dx.doi.org/ 10.1016/j.abb.2008.10.034 [DOI] [PubMed] [Google Scholar]
- 2.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc Nat Acad Sci USA 2004; 101:4003-8; PMID:15020765; http://dx.doi.org/ 10.1073/pnas.0307446101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corpas FJ, Leterrier M, Begara-Morales JC, Valderrama R, Chaki M, López-Jaramillo J, Luque F, Palma JM, Padilla MN, Sánchez-Calvo B, et al.. Inhibition of peroxisomal hydroxypyruvate reductase (HPR1) by tyrosine nitration. Biochim Biophys Acta 2013; 1830:4981-9; PMID:23860243; http://dx.doi.org/ 10.1016/j.bbagen.2013.07.002 [DOI] [PubMed] [Google Scholar]
- 4.Corpas FJ, Palma JM, Del Río LA. Protein tyrosine nitration in higher plants grown under natural and stress conditions. Front Plant Sci 2013; 4:29; PMID:23444154; http://dx.doi.org/ 10.3389/fpls.2013.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yadav S, David A, Baluška F, Bhatla SC. Rapid auxin-induced nitric oxide accumulation and subsequent tyrosine nitration of proteins during adventitious root formation in sunflower hypocotyls. Plant Signal Behav 2013; 8(3):e23196; PMID:23299324; http://dx.doi.org/ 10.4161/psb.23196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cecconi D, Orzetti S, Vandelle E, Rinalducci S, Zolla L, Delledonne M. Protein nitration during defense response in Arabidopsis thaliana. Electrophoresis 2009; 30:2460-68; PMID:19598157; http://dx.doi.org/ 10.1002/elps.200800826 [DOI] [PubMed] [Google Scholar]
- 7.Galetskiy D, Lohscheider JN, Kononikhin AS, Popov IA, Nikolaev EN, Adamska I. Mass spectrometric characterization of photooxidative protein modifications in Arabidopsis thaliana thylakoid membranes. Rapid Commun Mass Spectrom 2011; 25:184-90; PMID:21154902; http://dx.doi.org/ 10.1002/rcm.4855 [DOI] [PubMed] [Google Scholar]
- 8.Lozano-Juste J, Colom-Moreno R, León J. In vivo protein tyrosine nitration in Arabidopsis thaliana. J Exp Bot 2011; 62:3501-17; PMID:21378116; http://dx.doi.org/ 10.1093/jxb/err042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi M, Shigeto J, Sakamoto A, Izumi S, Asada K, Morikawa H. Dual selective nitration in Arabidopsis: almost exclusive nitration of PsbO and PsbP, and highly susceptible nitration of four non-PSII proteins, including peroxiredoxin II E. Electrophoresis 2015; 36:2569-78; PMID:26177577; http://dx.doi.org/ 10.1002/elps.201500145 [DOI] [PubMed] [Google Scholar]
- 10.Chaki M, Valderrama R, Fernández-Ocaña AM, Carreras A, Gómez-Rodríguez MV, López-Jaramillo J, Begara-Morales JC, Sánchez-Calvo B, Luque F, Leterrier M, et al.. High temperature triggers the metabolism of S-nitrosothiols in sunflower mediating a process of nitrosative stress which provokes the inhibition of ferredoxin-NADP reductase by tyrosine nitration. Plant Cell Environ 2011; 34:1803-18; PMID:21676000; http://dx.doi.org/ 10.1111/j.1365-3040.2011.02376.x [DOI] [PubMed] [Google Scholar]
- 11.Chaki M, Valderrama R, Fernández-Ocaña AM, Carreras A, López-Jaramillo J, Luque F, Palma JM, Pedrajas JR, Begara-Morales JC, Sánchez-Calvo B, et al.. 2009. Protein targets of tyrosine nitration in sunflower (Helianthus annuus L.) hypocotyls. J Exp Bot 2009; 60:4221-34; PMID:19717529; http://dx.doi.org/ 10.1093/jxb/erp263 [DOI] [PubMed] [Google Scholar]
- 12.Begara-Morales JC, Chaki M, Sánchez-Calvo B, Mata-Pérez C, Leterrier M, Palma JM, Barroso JB, Corpas FJ. Protein tyrosine nitration in pea roots during development and senescence. J Exp Bot 2013; 64:1121-1134; PMID:23362300; http://dx.doi.org/ 10.1093/jxb/ert006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Görg A, Postel W, Günther S, Weser J. Improved horizontal two-dimensional electrophoresis with hybrid isoelectric focusing in immobilized pH gradients in the first dimension and laying-on transfer to the second dimension. Electrophoresis 1985; 6:599-604; http://dx.doi.org/ 10.1002/elps.1150061206 [DOI] [Google Scholar]
- 14.Takahashi M, Sasaki Y, Ida S, Morikawa H. Enrichment of nitrite reductase gene improves the ability of Arabidopsis thaliana plants to assimilate nitrogen dioxide. Plant Physiol 2001; 126:731-41; PMID:11402201; http://dx.doi.org/ 10.1104/pp.126.2.731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem 1996; 68:850-8; PMID:8779443; http://dx.doi.org/ 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- 16.Kristensen DB, Imamura K, Miyamoto Y, Yoshizato K. Mass spectrometric approaches for the characterization of proteins on a hybrid quadrupole time-of-flight (Q-TOF) mass spectrometer. Electrophoresis 2000; 21:430-9; PMID:10675024; http://dx.doi.org/ 10.1002/(SICI)1522-2683(20000101)21:2%3c430::AID-ELPS430%3e3.0.CO;2-0 [DOI] [PubMed] [Google Scholar]
- 17.Matsuo E1, Toda C, Watanabe M, Ojima N, Izumi S, Tanaka K, Tsunasawa S, Nishimura O. Selective detection of 2-nitrobenzenesulfenyl-labeled peptides by matrix-assisted laser desorption/ionization-time of flight mass spectrometry using a novel matrix. Proteomics 2006; 6:2042-9; PMID:16521152; http://dx.doi.org/ 10.1002/pmic.200500575 [DOI] [PubMed] [Google Scholar]
- 18.Edreva A. Pathogenesis-related proteins: research progress in the last 15 years. Gen App Plant Physiol 2005; 31:105-24. [Google Scholar]
- 19.van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Ann Rev Phytopathol 2006; 44:135-62; PMID:16602946; http://dx.doi.org/24272264 10.1146/annurev.phyto.44.070505.143425 [DOI] [PubMed] [Google Scholar]
- 20.Payne G, Middlesteadt W, Williams S, Desai N, Parks TD, Dincher S, Carnes M, Ryals J. Isolation and nucleotide sequence of a novel cDNA clone encoding the major form of pathogenesis-related protein R. Plant Mol Biol 1988; 11:223-24; PMID:24272264; http://dx.doi.org/ 10.1007/BF00015674 [DOI] [PubMed] [Google Scholar]
- 21.Carr JP, Dixon DC, Nikolau BJ, Voelkerding KV, Klessig DF. Synthesis and localization of pathogenesis-related proteins in tobacco. Mol Cell Biol 1987; 7:1580-83; PMID:3299048; http://dx.doi.org/ 10.1128/MCB.7.4.1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon DC, Cutt JR, Klessig DF. Differential targeting of the tobacco PR-1 pathogenesis-related proteins to the extracellular space and vacuoles of crystal idioblasts. EMBO J 1991; 10:1317-24; PMID:2026137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park KS, Frost BF, Shin S, Park IK, Kim S, Paik WK. Effect of enzymatic methylation of yeast iso-1-cytochrome c on its isoelectric point. Arch Biochem Biophys 1988; 267:195-204; PMID:2848448; http://dx.doi.org/ 10.1016/0003-9861(88)90023-9 [DOI] [PubMed] [Google Scholar]
- 24.Jasid S, Simontacchi M, Bartoli CG, Puntarulo S. Chloroplasts as a nitric oxide cellular source. Effect of reactive nitrogen species on chloroplastic lipids and proteins. Plant Physiol 2006; 142:1246-55; PMID:16980561; http://dx.doi.org/ 10.1104/pp.106.086918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barroso JB, Valderrama R, Corpas FJ. Immunolocalization of S-nitrosoglutathione, S-nitrosoglutathione reductase and tyrosine nitration in pea leaf organelles. Acta Physiol Plant 2013; 35:2635-40; http://dx.doi.org/ 10.1007/s11738-013-1291-0 [DOI] [Google Scholar]
- 26.Durner J, Wendehenne D, Klessig DF. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Nat Acad Sci USA 1998; 95:10328-33; PMID:9707647; http://dx.doi.org/ 10.1073/pnas.95.17.10328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuoka M, Ohashi Y. Induction of pathogenesis-related proteins in tobacco leaves. Plant Physiol 1986; 80:505-10; PMID:16664652; http://dx.doi.org/ 10.1104/pp.80.2.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yalpani N, Enyedi AJ, León J, Raskin I. Ultraviolet light and ozone stimulate accumulation of salicylic acid, pathogenesis-related proteins and virus resistance in tobacco. Planta 1994; 193:372-6; http://dx.doi.org/ 10.1007/BF00201815 [DOI] [Google Scholar]
- 29.Takahashi M, Nakagawa M, Sakamoto A, Ohsumi C, Matsubara T, Morikawa H. Atmospheric nitrogen dioxide gas is a plant vitalization signal to increase plant size and the contents of cell constituents. New Phytol 2005; 168:149-54; PMID:16159329; http://dx.doi.org/ 10.1111/j.1469-8137.2005.01493.x [DOI] [PubMed] [Google Scholar]
- 30.Adam SEH, Shigeto J, Sakamoto A, Takahashi M, Morikawa H. Atmospheric nitrogen dioxide at ambient levels stimulates growth and development of horticultural plants. Botany 2008; 86:213-17; http://dx.doi.org/ 10.1139/B07-129 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


