Figure 4. miR-137 and miR-6500-3p target CENPE, KIF14 or NCAPG, which are upregulated in pHGGs.
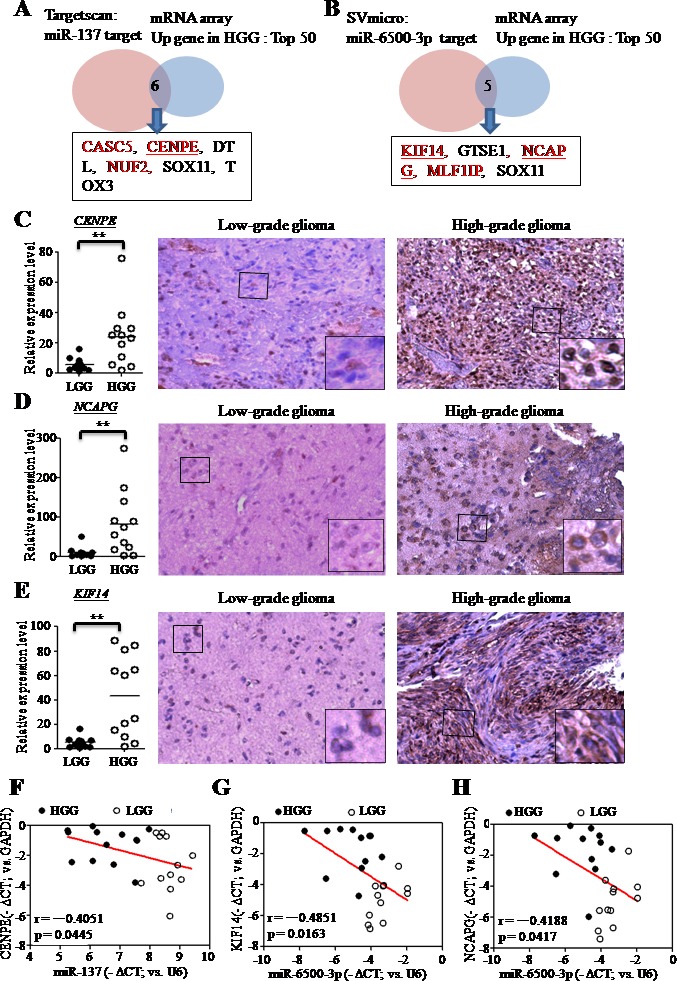
Venn diagram showing overlapping mRNAs based on microarray data (top 50 upregulated mRNAs in pHGGs) and bioinformatics prediction results (Targetscan or SVmicro). The bioinformatic analysis indicates the miR-137 A. and miR-6500-3p B. targets. RT-qPCR and IHC analyses confirmed higher mRNA and protein levels of CENPE C., KIF14 D. and NCAPG E. in pHGGs (mRNA: n = 12; protein: n = 3) than in pLGGs (mRNA: n = 12; protein: n = 3). RT-qPCR results are presented as mean±SD for duplicate samples. **p < 0.01 by t-test. Scatter plots illustrating the negative correlations between miR-137 and CENPE F., miR-6500-3p and KIF14 G., and miR-6500-3p and NCAPG H. in pediatric gliomas.
