Abstract
Forkhead box protein A1 (FOXA1) is a transcription factor; recent studies have reported that FOXA1 has an oncogenic or tumor suppressive role in human malignancies, and its expression is associated with the prognosis of patients with cancer. However, further studies are required to determine the clinical significance of FOXA1 and its role in colorectal cancer (CRC). In the present study, FOXA1 expression was detected in 90 samples of CRC tissues and matched noncancerous tissues using immunohistochemistry. In these cases, FOXA1 expression was detected in 57.8% (52/90) of the CRC samples, whereas only 37.8% (34/90) of the noncancerous specimens exhibited a positive FOXA1 signal. In addition, the present study demonstrated that the mRNA expression levels of FOXA1 were significantly increased in CRC tissues compared with in matched tumor-adjacent tissues. Furthermore, the positive expression of FOXA1 was associated with poor clinicopathological characteristics of CRC, including poor tumor differentiation, large tumor size, lymph node metastases and advanced tumor-node-metastasis tumor stage. Notably, patients with CRC with positive FOXA1 expression exhibited a significantly reduced 5-year survival rate compared with those with negative FOXA1 expression. Multivariate Cox regression analysis revealed that FOXA1 expression was an independent prognostic indicator for patients with CRC. In addition, FOXA1 knockdown evidently inhibited cell proliferation and induced apoptosis in SW480 and HCT116 CRC cells. Notably, FOXA1 knockdown also prominently reduced the expression of yes-associated protein (YAP) in SW480 and HCT116 cells. In conclusion, the results of the present study indicated that FOXA1 may be considered a potential prognostic marker, and may promote tumor growth of CRC by upregulating YAP expression.
Keywords: forkhead box protein A1, colorectal cancer, proliferation, apoptosis, yes-associated protein
Introduction
Colorectal cancer (CRC) is one of the most common types of malignancy worldwide, with an increasing incidence in China (1). In addition, it is the fourth leading cause of cancer-associated mortality, and is responsible for 529,000 cases of mortality per year worldwide (2). The long-term prognosis for patients with CRC remains poor, with a 50–59% 5-year survival rate (2). Early diagnosis and the use of molecular targeted therapies are effective at improving the clinical outcomes for patients with CRC. Therefore, it is important to disclose the molecular mechanisms underlying the initiation and development of CRC, which may provide evidence to explore novel biomarkers and therapeutic targets of CRC.
Forkhead box (FOX) protein A1 (FOXA1) is a pioneer transcription factor that belongs to the FOX gene superfamily (3) and exerts fundamental functions in the process of development and differentiation (4–8). FOXA1 modulates transcriptional programs in a tissue-dependent manner by inducing nucleosomal rearrangement and altering the accessibility of chromatin (3). In addition, FOXA1 is responsible for various biological processes, including cell proliferation, apoptosis and differentiation (9). Due to its critical role in these cellular processes, investigators have begun to determine its functional significance in human cancers. Elevated FOXA1 expression has been confirmed in anaplastic thyroid cancer (10), lung cancer (11,12) and hepatocellular carcinoma (HCC) (13), whereas decreased FOXA1 expression has been detected in poorly differentiated pancreatic cancer tissues (14). Functionally, FOXA1 is able to promote the proliferation of thyroid cancer cells (10), potentiate the metastatic ability of prostate cancer (15), and has been associated with sexual dimorphism of HCC (16). However, until now, the expression and biological function of FOXA1 in CRC have yet to be elucidated.
The present study aimed to determine the difference in FOXA1 expression between CRC tissues and noncancerous tissues. The clinical significance of FOXA1 was also explored. Furthermore, the role of FOXA1 in CRC growth and the potential underlying mechanisms were also addressed.
Materials and methods
Patients and clinicopathological data
A total of 90 samples of CRC and matched tumor-adjacent tissues were collected from the Department of Pathology, The Second Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China) between January 2007 and January 2009. The demographic features and clinicopathological parameters are presented in Table I. All specimens had a confirmed pathological diagnosis and were classified according to International Union Against Cancer and American Joint Committee on Cancer criteria (7th edition) (17). Patients did not receive preoperative chemotherapy, biotherapy or molecular targeted therapy. All samples were used after obtaining informed consent from the patients. The Xi'an Jiaotong University Ethics Committee approved all protocols, according to the Declaration of Helsinki (as revised in Tokyo, 2004).
Table I.
Clinical association analysis of FOXA1 expression in colorectal cancer.
| Feature | Total no. of patients, n=90 | No. of patients
|
P | |
|---|---|---|---|---|
| FOXA1 positive | FOXA1 negative | |||
| Age (years) | 0.094 | |||
| ≤60 | 27 | 12 | 15 | |
| >60 | 63 | 40 | 23 | |
| Gender | 0.223 | |||
| Male | 54 | 34 | 20 | |
| Female | 36 | 18 | 18 | |
| Tumor grade | 0.005a | |||
| G1/G2 | 67 | 33 | 34 | |
| G3/G4 | 23 | 19 | 4 | |
| Size (cm) | 0.004a | |||
| <5 | 32 | 12 | 20 | |
| ≥5 | 58 | 40 | 18 | |
| Tumor invasion | 0.458 | |||
| T1/T2 | 20 | 13 | 7 | |
| T3/T4 | 70 | 39 | 31 | |
| Lymph node metastases | 0.040a | |||
| Absent | 63 | 32 | 31 | |
| Present | 27 | 20 | 7 | |
| Distant metastasis | 0.609 | |||
| Absent | 71 | 42 | 29 | |
| Present | 19 | 10 | 9 | |
| TNM stage | 0.023a | |||
| I/II | 54 | 26 | 28 | |
| III/IV | 36 | 26 | 10 | |
Statistically significant. FOXA1, forkhead box protein A1; TNM, tumor-node-metastasis.
Immunohistochemical staining
The tissue of interest was fixed by immersing it in 10% neutral buffered formalin for 4–24 h at room temperature. The tissue was subsequently embedded in paraffin and the blocks were stored at 4°C prior to sectioning. The formalin-fixed and paraffin-embedded tissues from postoperative patients were cut into 5 µm sections. Sections were deparaffinized in xylene and rehydrated using a series of graded alcohols. Subsequently, tissue slides were subjected to antigen retrieval using 0.01 M sodium citrate (pH 6.0) at a sub-boiling temperature for 10 min and were blocked with 10% goat serum (ZSGB-Bio, Beijing, China). The samples were then incubated overnight at 4°C with anti-FOXA1 (1:100; cat. no. sc-101058; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) antibody. Immunostaining was performed using a SPlink Detection kit (cat. no. SP-9002; ZSGB-Bio) according to the manufacturer's protocol. Staining intensity was scored as follows: 0, negative; 1, weak; or 2, strong; and the extent of staining was based on the percentage of positive tumor cells: 0, negative; 1, 1–25%; 2, 26–50%; 3, 51–75%; or 4, 76–100% (18). Therefore, each case was ultimately considered negative if the final score was 0 (negative expression) or 1–2 (low expression), and positive if the final score was 3–5 (medium expression) or 6–8 (high expression). The final immunohistochemistry score for each sample (negative or positive) was calculated by multiplying the intensity score with the percentage of positive cells.
Cell culture and transfection
Two CRC cell lines, HCT116 and SW480 (The Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China), were purchased for use in the present study. Cells were routinely cultured in complete Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) at 37°C in a humidified incubator containing 5% CO2.
The targeted sequences for FOXA1 small interfering (si)RNA (sense 5′-GCACUGCAAUACUCGCCUU-3′) or a nonspecific duplex oligonucleotide as a negative control were synthesized by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). Cells were transfected with the aforementioned siRNAs using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol.
Immunoblotting
The tissue was snap-frozen in liquid nitrogen and diced into 1 mm pieces with a scalpel in a mortar on dry ice. Total proteins were extracted using radioimmunoprecipitation assay lysis buffer (Santa Cruz Biotechnology, Inc.) and the protein concentrations were measured using the bicinchoninic acid assay. The proteins (20 µg) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were transferred onto a polyvinylidene fluoride membrane (Roche Diagnostics, Indianapolis, IN, USA). The membrane was then blocked with 5% skimmed milk at room temperature for 15 min and incubated with the following appropriate primary antibodies at 4°C overnight: FOXA1 (1:1,000), yes-associated protein (YAP; 1:1,000; cat. no. PA1-46189; Thermo Fisher Scientific, Inc., Rockford, IL, USA), caspase-3 (1:1,000; cat. no. 9662; Cell Signaling Technology, Inc., Danvers, MA, USA) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 1:5,000; cat. no. sc-25778; Santa Cruz Biotechnology, Inc.). Blots were then incubated with horseradish peroxidase-conjugated goat anti-mouse/rabbit immunoglobulin G secondary antibodies (1:5,000; cat. nos. CW0102/103; Cwbiotech, Shanghai, China) at room temperature for 2 h. Blots were detected using enhanced chemiluminescence regents (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and semi-quantified by ImageJ software (1.46; National Institutes of Health, Bethesda, MD, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used for total RNA extraction following tissue homogenization. cDNA synthesis was performed using PrimeScript RT Reagent kit (Takara Bio, Inc., Otsu, Japan) at 37°C for 15 min, following treatment with DNase I (Invitrogen; Thermo Fisher Scientific, Inc.). Amplification and detection of FOXA1 and GAPDH mRNA were performed using an ABI PRISM 7300 Sequence Detection system (Applied Biosystems, Inc., Foster City, CA, USA) and a One Step SYBR® PrimeScript™ RT-PCR kit II (Takara Bio, Inc.) according to the manufacturers' protocols. The following primers (Sangon Biotech (Shanghai) Co., Ltd.) were used: FOXA1, sense primer 5′-AAT CATTGCCATCGTGTG-3′, antisense primer 5′-CGCGGCTTAAAATCTGGTAT-3′; and GAPDH, sense 5′-CAAGCTCATTTCCTGGTATGAC-3′, and antisense 5′-CAGTGAGGGTCTCTCTCTTCCT-3′. The thermocycling conditions were as follows: 95°C for 60 sec; followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec. GAPDH was measured as an internal control for mRNA. All samples were normalized to internal controls and fold changes were calculated based on relative quantification using the 2−ΔΔCq method (13).
Proliferation and apoptosis assay
For the proliferation assay, CRC cells were seeded into 96-well plates at a density of 5,000 cells/well for 24 h and were assessed using Cell Proliferation enzyme-linked immunosorbent assay, 5-bromodeoxyuridine (BrdU) (chemiluminescent) (Roche Diagnostics) according to the manufacturer's protocol. Flow cytometric analysis was conducted using fluorescence-activated cell sorting (FACS)Calibur (BD Biosciences, San Jose, CA, USA) and Cell Quest Pro software (BD Biosciences). An Annexin-V-FLUOS Staining kit (Roche Diagnostics) was used to analyze the level of apoptosis according to the manufacturer's protocol. The percentage of apoptotic cells was calculated using the flow cytometry software.
Statistical analysis
Results are expressed as the mean ± standard error of the mean. All experiments were repeated three times. All statistical analyses were performed using SPSS statistical package for Windows version 13 (SPSS, Inc., Chicago, IL, USA) or GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). The quantitative data were compared between groups using the Student's t-test. Categorical data were analyzed using the Pearson chi-squared test. The Kaplan-Meier method and log-rank test were used to compare the cumulative recurrence and survival rates. The independent factors influencing the survival and recurrence of patients with CRC were determined using the Cox proportional hazards model. Correlation analysis was conducted using the Pearson's correlation coefficient. P<0.05 was considered to indicate a statistically significant difference.
Results
FOXA1 expression is elevated in CRC tissues
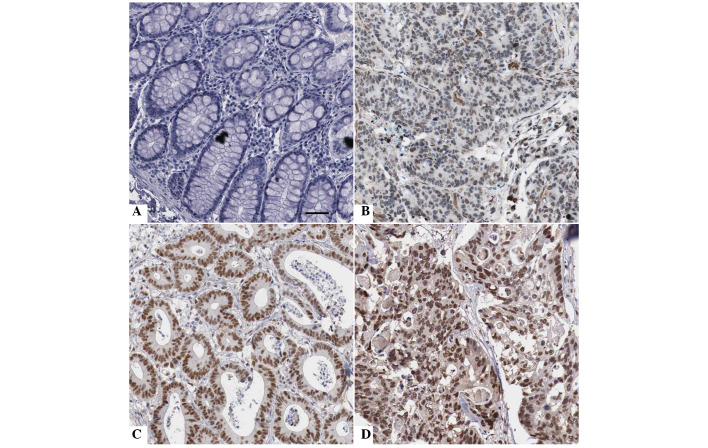
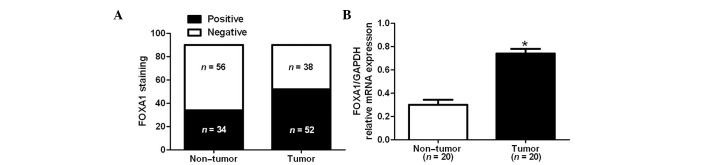
Immunohistochemical staining was performed to investigate differences in FOXA1 protein expression between CRC and adjacent noncancerous tissues. As shown in Fig. 1, FOXA1 staining was negative in the adjacent noncancerous tissue (Fig. 1A), whereas positive FOXA1 staining with nuclear location was observed in CRC tissues (Fig. 1B–D). FOXA1 expression was detected in 57.8% (52/90) of the CRC samples, whereas only 37.8% (34/90) of the noncancerous specimens exhibited a positive FOXA1 signal (P<0.05; Fig. 2A). Furthermore, 20 randomly selected cases were subjected to RT-qPCR in order to determine FOXA1 mRNA expression. The mRNA expression levels of FOXA1 were significantly increased in CRC tissues compared with in the adjacent noncancerous tissues (P<0.05; Fig. 2B). There results indicate an oncogenic role for FOXA1 in CRC.
Figure 1.
Immunostaining of forkhead box protein A1 (FOXA1) in colorectal cancer (CRC) and tumor-adjacent tissues. (A) Negative staining of FOXA1 in tumor-adjacent tissues; (B) low, (C) medium and (D) high expression of FOXA1 in CRC tissues. Scale bar: 50 µm.
Figure 2.
Differences in the expression of forkhead box protein A1 (FOXA1) in colorectal cancer (CRC) and tumor-adjacent tissues. (A) Immunohistochemical staining analysis indicated that the positive expression rate of FOXA1 in CRC tissues (52/90) was significantly higher than in adjacent non-tumor tissues (34/90). (B) Reverse transcription-quantitative polymerase chain reaction demonstrated that the mRNA expression levels of FOXA1 were significantly increased in CRC tissues compared with in adjacent non-tumor tissues. *P<0.05, as determined by Student's t-test. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Positive expression of FOXA1 is associated with poor clinicopathological features
To elucidate the clinical significance of FOXA1 expression in CRC, the present study investigated the association between FOXA1 expression and clinicopathological features of patients with CRC. As shown in Table I, positive FOXA1 expression in CRC tissues was associated with poor tumor differentiation (P=0.005), large tumor size (P=0.004), lymph node metastasis (P=0.040) and advanced tumor-node-metastasis stage (P=0.023). These results indicate that FOXA1 may promote the development and progression of CRC.
FOXA1 is a prognostic predictor for patients with CRC
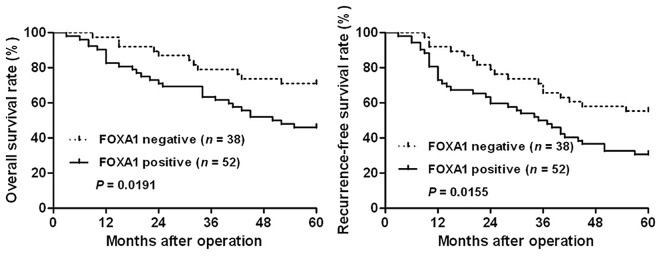
To further investigate the prognostic value of FOXA1 expression, the overall survival (OS) and recurrence-free survival (RFS) rates were compared between the FOXA1-positive group (n=52) and the FOXA1-negative group (n=38). Kaplan-Meier survival curves indicated that the positive expression of FOXA1 in CRC was significantly correlated with reduced OS (P=0.0191; Fig. 3) rate and RFS rates (P=0.0155; Fig. 3). Notably, a multivariate Cox regression analysis demonstrated that FOXA1 expression was an independent factor for predicting the OS and RFS of patients with CRC (P=0.005 and 0.017, respectively; Table II). These data indicate that FOXA1 expression is a promising predictor for patient prognosis in CRC.
Figure 3.
Prognostic significance of forkhead box protein A1 (FOXA1) for patients with colorectal cancer (CRC). Patients with CRC were divided into two groups according to FOXA1 immunostaining: FOXA1-positive group (n=52) and FOXA1-negative group (n=38). Kaplan-Meier plots indicated that the overall survival and recurrence-free survival rates were significantly reduced in the FOXA1-positive group compared with in the FOXA1-negative group.
Table II.
Multivariate Cox regression analysis of 5-year overall and recurrence-free survival rates of 90 patients with colorectal cancer.
| Variable | Overall survival
|
Recurrence-free survival
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Tumor grade (G1/G2 vs. G3/G4) | 1.904 | 0.734–4.982 | 0.190 | 2.343 | 0.950–5.812 | 0.065 |
| Tumor size (cm) (<5 vs. ≥5) | 1.233 | 0.650–2.023 | 0.342 | 1.371 | 0.864–2.561 | 0.426 |
| Lymph node metastases (absent vs. present) | 1.403 | 0.703–2.783 | 0.345 | 1.125 | 0.634–2.003 | 0.692 |
| TNM stage (I/II vs. III/IV) | 2.895 | 1.213–6.695 | 0.034a | 2.197 | 1.148–4.291 | 0.023a |
| FOXA1 expression (negative vs. positive) | 2.028 | 1.235–3.317 | 0.005a | 2.023 | 1.134–3.603 | 0.017a |
Statistically significant. FOXA1, forkhead box protein A1; TNM, tumor-node-metastasis; HR, hazard ratio; CI, confidence interval.
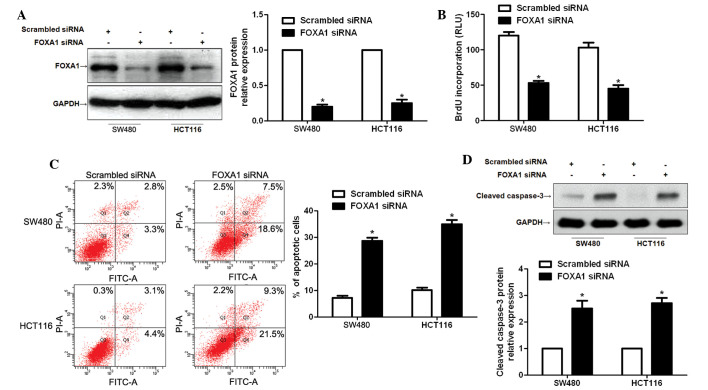
FOXA1 knockdown inhibits CRC cell proliferation and promotes apoptosis in vitro
To determine the functional role of FOXA1 in CRC, FOXA1 siRNA was used to inhibit the expression of FOXA1 in SW480 and HCT116 cells, and the proliferation and apoptosis of CRC cells was subsequently investigated. FOXA1 knockdown was confirmed by immunoblotting (P<0.05; Fig. 4A). Subsequently, BrdU incorporation assays demonstrated that the proliferation of SW480 and HCT116 cells was significantly decreased following FOXA1 knockdown (P<0.05; Fig. 4B). In addition, the percentage of apoptotic SW480 and HCT116 cells was significantly increased following suppression of FOXA1 (P<0.05; Fig. 4C). Western blot analysis indicated that FOXA1 knockdown evidently increased the protein expression of cleaved caspase-3 in SW480 and HCT116 cells (P<0.05; Fig. 4D).
Figure 4.
Knockdown of forkhead box protein A1 (FOXA1) suppresses colorectal cancer cell proliferation and promotes apoptosis. (A) SW480 and HCT116 cells were transfected with FOXA1 small interfering (si)RNA or scrambled siRNA, and were subjected to western blotting to detect FOXA1 expression. n=3 independent experiments. *P<0.05, as determined by Student's t-test. (B) FOXA1 knockdown significantly inhibited cell proliferation as measured by 5-bromodeoxyuridine (BrdU) incorporation in SW480 and HCT116 cells. n=3 independent experiments. *P<0.05, as determined by Student's t-test. (C) Apoptosis of SW480 and HCT116 cells was measured by flow cytometry, and was promoted by FOXA1 knockdown. n=3 independent repeats with similar results. *P<0.05, as determined by Student's t-test. (D) Western blot analysis indicated that FOXA1 knockdown increased the protein expression of cleaved caspase-3 in SW480 and HCT116 cells. n=3 independent experiments. *P<0.05, as determined by Student's t-test. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; RLU, relative light units; FITC, fluorescein isothiocyanate; PI, propidium iodide.
FOXA1 knockdown inhibits YAP expression in CRC cells
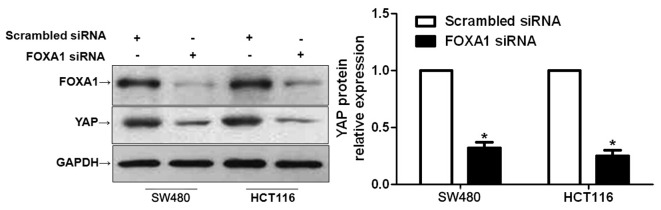
Hippo-YAP signaling has been confirmed to have a fundamental role in the pathogenesis of CRC (19,20). Furthermore, inhibiting YAP expression has been reported to lead to a marked decrease in the proliferation and apoptosis of CRC cells (21). A recent study reported that FOXA1 could regulate YAP expression in HCC cells (13). Therefore, the present study investigated whether the functional effects of FOXA1 in CRC cells were mediated via the modulation of YAP expression. As expected, FOXA1 knockdown resulted in a significant decrease in YAP expression in SW480 and HCT116 cells (P<0.05; Fig. 5). These data indicate that FOXA1 may regulate YAP expression in CRC cells.
Figure 5.
Downregulation of forkhead box protein A1 (FOXA1) reduces the expression of yes-associated protein (YAP) in colorectal cancer cells. SW480 and HCT116 cells were transfected with FOXA1 small interfering (si)RNA or scrambled siRNA, and were subjected to western blotting to determine YAP expression. Semi-quantitative analysis revealed that FOXA1 knockdown significantly reduced the protein expression levels of YAP. n=3 independent repeats with similar results. *P<0.05, as determined by Student's t-test. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Discussion
The FOXA family [also known as the hepatocyte nuclear factor 3 (HNF3) family] consists of FOXA1 (HNF3α), FOXA2 (HNF3β) and FOXA3 (HNF3γ). It is well established that dysregulation of FOXA members has a pivotal role in carcinogenesis (22). FOXA1 is a key transcription factor of the FOXA family, which exhibits aberrant expression and functions as a prognostic indicator in various types of human cancer (13,23,24). The present study was the first, to the best of our knowledge, to investigate the expression status of FOXA1 in CRC. Significantly elevated mRNA and protein expression of FOXA1 was detected in CRC tissues compared with in the tumor-adjacent tissues. A clinical association analysis demonstrated that FOXA1 expression was correlated with poor clinicopathological features and reduced survival of patients with CRC. In addition, a Cox regression analysis revealed that the positive expression of FOXA1 may be a potential factor for predicting a bad patient outcome.
A previous study reported that FOXA1 knockdown inhibited the effects of ELL-associated factor 2 knockdown, and was associated with decreased cell proliferation and migration in LNCaP cells (25). Qiu et al demonstrated that FOXA1 knockdown reduced the tumor growth of endometrial cancer in vivo and in vitro (26). Furthermore, FOXA1 has been reported to promote cell proliferation and inhibit apoptosis in HCC cells (13). All these studies prompted the investigation of the biological role of FOXA1 in CRC cells in the present study. The present in vitro experiments demonstrated that FOXA1 knockdown inhibited proliferation and induced apoptosis of CRC cells. These results indicated that FOXA1 may function as an oncogenic factor by promoting proliferation and suppressing apoptosis of CRC cells. The Hippo pathway has been revealed to be an important regulator of CRC cell proliferation, growth and apoptosis (19–21,27). The majority of genes coding for Hippo pathway proteins have been shown to function in several types of cancer, either as tumor suppressors or as oncogenes (28). The most commonly focused gene, YAP, has been reported to be involved in tumor development and progression in various malignancies, including CRC (28). YAP has been found to be significantly upregulated in CRC tissues, and affects the proliferation and apoptosis of CRC cells (21,29,30). In addition, it has been considered a therapeutic target of CRC (31). Notably, FOXA1 has a positive effect on YAP expression; opening of compacted chromatin by FOXA1 around the cAMP response element binding protein (CREB) binding site within the YAP promoter facilitates CREB-mediated YAP transcription in HCC cells (32). In the present study, FOXA1 expression was knocked down following transfection of CRC cells with a specific siRNA. Western blotting indicated that the protein expression levels of YAP were significantly decreased following FOXA1 knockdown. These results suggested that FOXA1 may regulate cell proliferation and apoptosis at least in part via the modulation of YAP expression in CRC cells.
In conclusion, the present study demonstrated that FOXA1 is overexpressed in CRC tissue specimens. The positive expression of FOXA1 was associated with poor clinicopathological features of CRC, thus suggesting that FOXA1 expression may be an independent indicator of prognosis in patients with CRC. Furthermore, FOXA1 may facilitate CRC cell proliferation and inhibit apoptosis by regulating YAP expression. Therefore, FOXA1 may be considered a potential prognostic biomarker and therapeutic target for CRC.
Acknowledgments
The present study was supported by a grant from the National Natural Science Foundation of China (grant no. 81171356).
References
- 1.Li L, Ma BB. Colorectal cancer in Chinese patients: Current and emerging treatment options. Onco Targets Ther. 2014;7:1817–1828. doi: 10.2147/OTT.S48409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Bernardo GM, Keri RA. FOXA1: A transcription factor with parallel functions in development and cancer. Biosci Rep. 2012;32:113–130. doi: 10.1042/BSR20110046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Augello MA, Hickey TE, Knudsen KE. FOXA1: Master of steroid receptor function in cancer. EMBO J. 2011;30:3885–3894. doi: 10.1038/emboj.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328:198–206. doi: 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Strazzabosco M. Foxa1 and Foxa2 regulate bile duct development in mice. J Hepatol. 2010;52:765–767. doi: 10.1016/j.jhep.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z, Merkurjev D, Yang F, Li W, Oh S, Friedman MJ, Song X, Zhang F, Ma Q, Ohgi KA, et al. Enhancer activation requires trans-recruitment of a mega transcription factor complex. Cell. 2014;159:358–373. doi: 10.1016/j.cell.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaret KS, Carroll JS. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nucera C, Eeckhoute J, Finn S, Carroll JS, Ligon AH, Priolo C, Fadda G, Toner M, Sheils O, Attard M, et al. FOXA1 is a potential oncogene in anaplastic thyroid carcinoma. Clin Cancer Res. 2009;15:3680–3689. doi: 10.1158/1078-0432.CCR-08-3155. [DOI] [PubMed] [Google Scholar]
- 11.Lin L, Miller CT, Contreras JI, Prescott MS, Dagenais SL, Wu R, Yee J, Orringer MB, Misek DE, Hanash SM, et al. The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1), on chromosome band 14q13 is amplified and overexpressed in esophageal and lung adenocarcinomas. Cancer Res. 2002;62:5273–5279. [PubMed] [Google Scholar]
- 12.Deutsch L, Wrage M, Koops S, Glatzel M, Uzunoglu FG, Kutup A, Hinsch A, Sauter G, Izbicki JR, Pantel K, Wikman H. Opposite roles of FOXA1 and NKX2-1 in lung cancer progression. Genes Chromosomes Cancer. 2012;51:618–629. doi: 10.1002/gcc.21950. [DOI] [PubMed] [Google Scholar]
- 13.Dou C, Wang Y, Li C, Liu Z, Jia Y, Li Q, Yang W, Yao Y, Liu Q, Tu K. MicroRNA-212 suppresses tumor growth of human hepatocellular carcinoma by targeting FOXA1. Oncotarget. 2015;6:13216–13228. doi: 10.18632/oncotarget.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70:2115–2125. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerhardt J, Montani M, Wild P, Beer M, Huber F, Hermanns T, Müntener M, Kristiansen G. FOXA1 promotes tumor progression in prostate cancer and represents a novel hallmark of castration-resistant prostate cancer. Am J Pathol. 2012;180:848–861. doi: 10.1016/j.ajpath.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148:72–83. doi: 10.1016/j.cell.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong LL, Gao P, Wang ZN, Song YX, Xu YY, Sun Z, Xing CZ, Xu HM. Is the seventh edition of the UICC/AJCC TNM staging system reasonable for patients with tumor deposits in colorectal cancer? Ann Surg. 2012;255:208–213. doi: 10.1097/SLA.0b013e31821ad8a2. [DOI] [PubMed] [Google Scholar]
- 18.Huang W, Chen Z, Shang X, Tian D, Wang D, Wu K, Fan D, Xia L. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology. 2015;61:1920–1933. doi: 10.1002/hep.27756. [DOI] [PubMed] [Google Scholar]
- 19.Avruch J, Zhou D, Bardeesy N. YAP oncogene overexpression supercharges colon cancer proliferation. Cell Cycle. 2012;11:1090–1096. doi: 10.4161/cc.11.6.19453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barry ER, Morikawa T, Butler BL, Shrestha K, de la Rosa R, Yan KS, Fuchs CS, Magness ST, Smits R, Ogino S, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Shi S, Guo Z, Zhang X, Han S, Yang A, Wen W, Zhu Q. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PLoS One. 2013;8:e65539. doi: 10.1371/journal.pone.0065539. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Zhao Y, Li Z. Interplay of estrogen receptors and FOXA factors in the liver cancer. Mol Cell Endocrinol. 2015;418:334–339. doi: 10.1016/j.mce.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy OL, Cates JM, Gellert LL, Crist HS, Yang Z, Yamashita H, Taylor JA, III, Smith JA, Jr, Chang SS, Cookson MS, et al. Loss of FOXA1 drives sexually dimorphic changes in urothelial differentiation and is an independent predictor of poor prognosis in bladder cancer. Am J Pathol. 2015;185:1385–1395. doi: 10.1016/j.ajpath.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horimoto Y, Arakawa A, Harada-Shoji N, Sonoue H, Yoshida Y, Himuro T, Igari F, Tokuda E, Mamat O, Tanabe M, et al. Low FOXA1 expression predicts good response to neo-adjuvant chemotherapy resulting in good outcomes for luminal HER2-negative breast cancer cases. Br J Cancer. 2015;112:345–351. doi: 10.1038/bjc.2014.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo W, Keener AL, Jing Y, Cai L, Ai J, Zhang J, Fisher AL, Fu G, Wang Z. FOXA1 modulates EAF2 regulation of AR transcriptional activity, cell proliferation, and migration in prostate cancer cells. Prostate. 2015;75:976–987. doi: 10.1002/pros.22982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu M, Bao W, Wang J, Yang T, He X, Liao Y, Wan X. FOXA1 promotes tumor cell proliferation through AR involving the Notch pathway in endometrial cancer. BMC Cancer. 2014;14:78. doi: 10.1186/1471-2407-14-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 28.Wierzbicki PM, Rybarczyk A. The Hippo pathway in colorectal cancer. Folia Histochem Cytobiol. 2015;53:105–119. doi: 10.5603/FHC.a2015.0015. [DOI] [PubMed] [Google Scholar]
- 29.Konsavage WM, Jr, Kyler SL, Rennoll SA, Jin G, Yochum GS. Wnt/β-catenin signaling regulates Yes-associated protein (YAP) gene expression in colorectal carcinoma cells. J Biol Chem. 2012;287:11730–11739. doi: 10.1074/jbc.M111.327767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Xie C, Li Q, Xu K, Wang E. Clinical and prognostic significance of Yes-associated protein in colorectal cancer. Tumour Biol. 2013;34:2169–2174. doi: 10.1007/s13277-013-0751-x. [DOI] [PubMed] [Google Scholar]
- 31.Liang K, Zhou G, Zhang Q, Li J, Zhang C. Expression of hippo pathway in colorectal cancer. Saudi J Gastroenterol. 2014;20:188–194. doi: 10.4103/1319-3767.133025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu W, Qiao Y, Tang X, Ma L, Wang Y, Zhang X, Weng W, Pan Q, Yu Y, Sun F, Wang J. Tumor suppressor long non-coding RNA, MT1DP is negatively regulated by YAP and Runx2 to inhibit FoxA1 in liver cancer cells. Cell Signal. 2014;26:2961–2968. doi: 10.1016/j.cellsig.2014.09.011. [DOI] [PubMed] [Google Scholar]