Abstract
Mutations of the neuronal sodium channel gene SCN8A are associated with lethal movement disorders in the mouse and with human epileptic encephalopathy. We describe a spontaneous mouse mutation, Scn8a 9J, that is associated with a chronic movement disorder with early onset tremor and adult onset dystonia. Scn8a 9J homozygotes have a shortened lifespan, with only 50% of mutants surviving beyond 6 months of age. The 3 bp in-frame deletion removes 1 of the 3 adjacent isoleucine residues in transmembrane segment DIVS6 of Nav1.6 (p.Ile1750del). The altered helical orientation of the transmembrane segment displaces pore-lining amino acids with important roles in channel activation and inactivation. The predicted impact on channel activity was confirmed by analysis of cerebellar Purkinje neurons from mutant mice, which lack spontaneous and induced repetitive firing. In a heterologous expression system, the activity of the mutant channel was below the threshold for detection. Observations of decreased nerve conduction velocity and impaired behavior in an open field are also consistent with reduced activity of Nav1.6. The Nav1.6Δ1750 protein is only partially glycosylated. The abundance of mutant Nav1.6 is reduced at nodes of Ranvier and is not detectable at the axon initial segment. Despite a severe reduction in channel activity, the lifespan and motor function of Scn8a9J/9J mice are significantly better than null mutants lacking channel protein. The clinical phenotype of this severe hypomorphic mutant expands the spectrum of Scn8a disease to include a recessively inherited, chronic and progressive movement disorder.
Keywords: voltage-gated sodium channel, movement disorder, transmembrane segment, glycosylation, axon initial segment, nodes of Ranvier, Purkinje cell, Scn8a, Nav1.6
INTRODUCTION
The sodium channel gene Scn8a encodes Nav1.6, an abundant voltage-gated sodium channel in mammalian brain and peripheral nerves. Within neurons, Nav1.6 is present at low levels in dendrites and soma and at high concentrations at the axon initial segment (AIS) and nodes of Ranvier (Caldwell et al., 2000; Lorincz and Nusser, 2008; Lorincz and Nusser, 2010). Nav1.6 has a lower threshold for generation of action potentials than Nav1.1 and Nav1.2, the other major brain channels (Spampanato et al., 2001; Rush et al., 2005; Chen et al., 2008), and is an important source of persistent and resurgent sodium current (Raman et al., 1997; Royeck et al., 2008; O'Brien and Meisler, 2013). These properties of Nav1.6 contribute to the generation of bursts of action potentials in repetitively firing neurons such as Purkinje cells (Raman et al., 1997; Lorincz and Nusser, 2008; Cruz et al., 2011). Nav1.6 is the major sodium channel at the AIS of cerebellar Purkinje cells, repetitively firing neurons that exhibit bursts of action potentials (Lorincz and Nusser, 2008).
Several null mutations with complete loss of Nav1.6 protein have been studied in the mouse (O'Brien and Meisler, 2013). Homozygous null mice develop hind limb paralysis by two weeks of age, when Nav1.6 replaces Nav1.1 and Nav1.2 at maturing nodes of Ranvier (Boiko et al., 2001) and juvenile lethality at 3 weeks of age (Burgess et al., 1995; Sharkey et al., 2009). The loss of innervation at the neuromuscular junction in the null mice results in nerve terminal sprouting and muscle atrophy (Duchen, 1970; Sprunger et al., 1999).
De novo missense mutations of human SCN8A are one cause of early infantile epileptic encephalopathy (OMIM# 614558) (Veeramah et al., 2012; Carvill et al., 2013; Estacion et al., 2014; Blanchard et al., 2015; Larsen et al., 2015; Wagnon and Meisler, 2015). Many of these mutations cause elevated neuronal excitability due to impaired channel inactivation (Veeramah et al., 2012, Wagnon et al., 2015) or hyperpolarizing shifts in voltage dependence of activation (Estacion et al., 2014; Blanchard et al., 2015). Approximately half of affected children are non-ambulatory and the majority of affected individuals speak few words. Intellectual disability ranges from mild to severe, with about half of affected individuals having severe intellectual disability (Larsen et al., 2015; Wagnon and Meisler, 2015).
The sodium channel pore-forming α subunit contains four homologous protein domains, DI to DIV, each with six transmembrane segments, S1 to S6 (Catterall et al., 2007). Site directed mutagenesis and the crystal structure of the bacterial sodium channel support a model in which transmembrane segments S1-S4 constitute the voltage-sensing domain of the channel, and segments S5 and S6 line the sodium-permeable pore (Payandeh et al., 2011). Sodium channel α subunits are N-glycosylated and contain terminal sialic acid residues. Core-glycosylation of the 228 kDa Nav1.2 protein in the endoplasmic reticulum and subsequent modification in the Golgi generate a 260 kDa fully glycosylated protein (Schmidt and Catterall, 1986). Nav1.6 from mammalian brain migrates with a MW similar to fully glycosylated Nav1.2 (Kearney et al., 2002; Wagnon et al., 2015). Glycosylation of the related channels Nav1.2, Nav1.4, Nav1.5 and Nav1.7 is known to influence protein folding, protease resistance, cell surface localization and channel kinetics (Ednie and Bennett, 2012; Baycin-Hizal et al., 2014; Lazniewska and Weiss, 2014). Core-glycosylated Nav1.5 and Nav1.7 can reach the surface of transfected HEK cells but may lack channel activity (Laedermann et al., 2013; Mercier et al., 2015).
We describe a new spontaneous mutation of Scn8a with unusual properties. The mutant protein is partially glycosylated and channel activity in endogenous neurons is greatly reduced. Homozygous mutant mice have an abnormal gait, progressively impaired movement, and reduced lifespan. The survival of this mutant is much longer than true null mutants lacking channel protein, consistent with a low level of residual activity. The new mouse model will be useful for understanding the in vivo function of Nav1.6 and the pathogenic mechanisms associated with patient mutations.
MATERIALS AND METHODS
Mice
The Scn8a9J mutation arose spontaneously in strain BALB/cJ at the Jackson Laboratory. The mutation was initially maintained by transfer of ovarian tissue from homozygous affected females to wildtype carriers. Scn8a9J/+ mice were subsequently backcrossed to strain C57BL/6J for 10 generations at the University of Michigan. Mice were housed and cared for in accordance with NIH guidelines. Experiments were approved by the University of Michigan Committee on the Use and Care of Animals. All experiments were carried out in homozygous mutant mice with the exception of open field behavior, which was assessed in heterozygous mutants. The controls were wildtype littermates unless otherwise stated.
Mutation identification
The 28 coding exons and intron/exon boundaries of Scn8a were amplified from genomic DNA isolated from affected mice. Gel-purified PCR products were analyzed by automated Sanger sequencing in the University of Michigan Sequencing Core.
Genotyping by RFLP-PCR
The 3 base pair deletion in the final coding exon of the Scn8a9J allele introduces a novel BsaXI restriction site. Mutant mice are identified by amplification of a 308 bp genomic fragment containing the mutation using the forward primer 9JF (5’ ATTGA GCGTC TGCAC GGTAA GAGG) and reverse primer 9JR (5’GTGGC ATCAG GATCA AACTT CTCC). PCR is carried out in a 25 µl volume containing 1X Coral Load DNA polymerase buffer (Qiagen), 2.5 mM MgCl2, 0.2 mM dNTPs, 0.5 uM primers and 1 unit DNA polymerase (Qiagen), with initial incubation for 3 min at 94°C followed by 31 cycles with 0.5 min at 94°C, 1 min at 66°C and 1 min at 72°C, ending with 6 min at 72°C. Digestion of the PCR product with BsaXI (New England Biolabs) generates fragments of 142 bp, 130 bp and 33 bp from the mutant allele and the intact 308 bp fragment from the wildtype allele. Restriction fragments are detected by electrophoresis on 2% agarose gels after staining with ethidium bromide.
Glycosylation and Western blots
Membrane proteins were prepared by centrifugation of brain homogenates at 100,000 x g as previously described (O'Brien et al., 2012). Aliquots containing 20 to 40 µg of protein were resuspended in 1 X glycoprotein denaturing buffer at room temperature and then incubated with 1500 units of PNGaseF1, 1x NP-40 and 1X G7 buffer (New England Biolabs) for 1 hour at 37°C. The reaction was stopped by addition of 1X sample buffer containing 50 mM Tris, pH 6.8, 1% SDS, 0.001% bromophenol blue, 9% glycerol and 1.8% 2-mercapoethanol. Products were examined by Western blotting as previously described (O'Brien et al., 2012). Blots were immunostained with polyclonal anti-Nav1.6 antiserum (Alomone Labs #ASC-009, 1:200 dilution), anti-human α-tubulin (Cedarlane Laboratories #CLT9002, 1:5,000 dilution), and antibody to the ubiquitously expressed lipid phosphatase FIG 4, a housekeeping enzyme, anti-mouse Fig 4 (Antibodies Incorporated #75–201, 1:100 dilution).
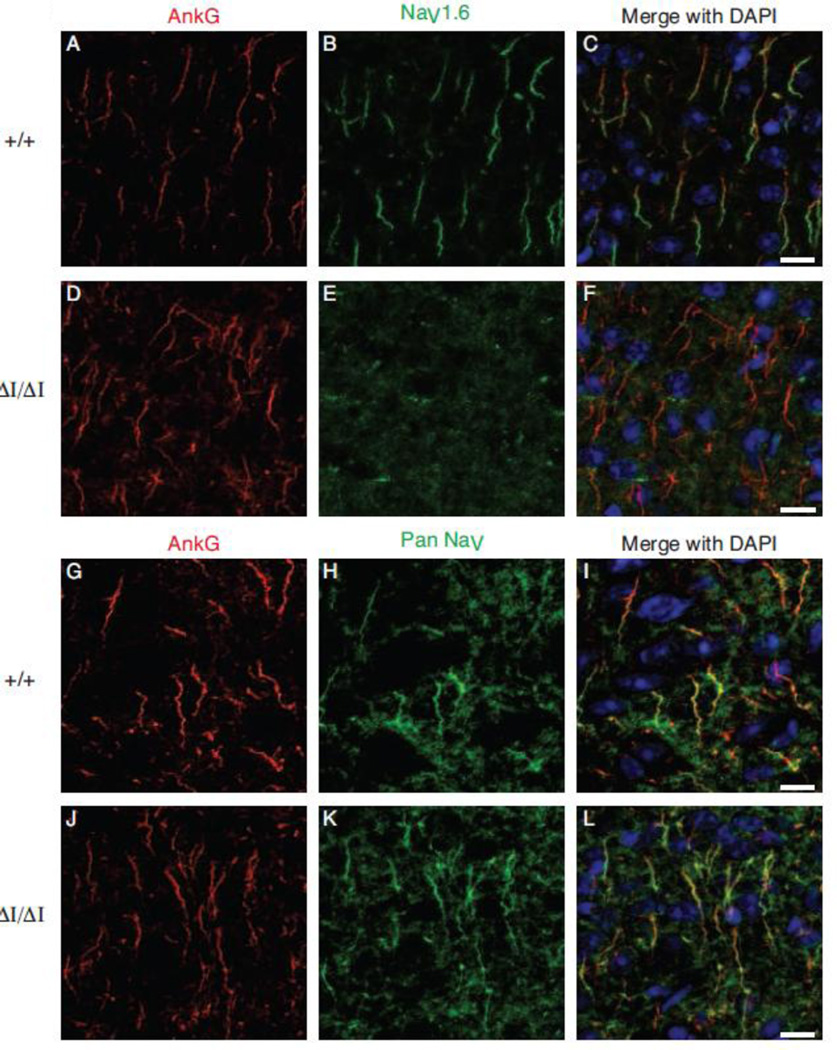
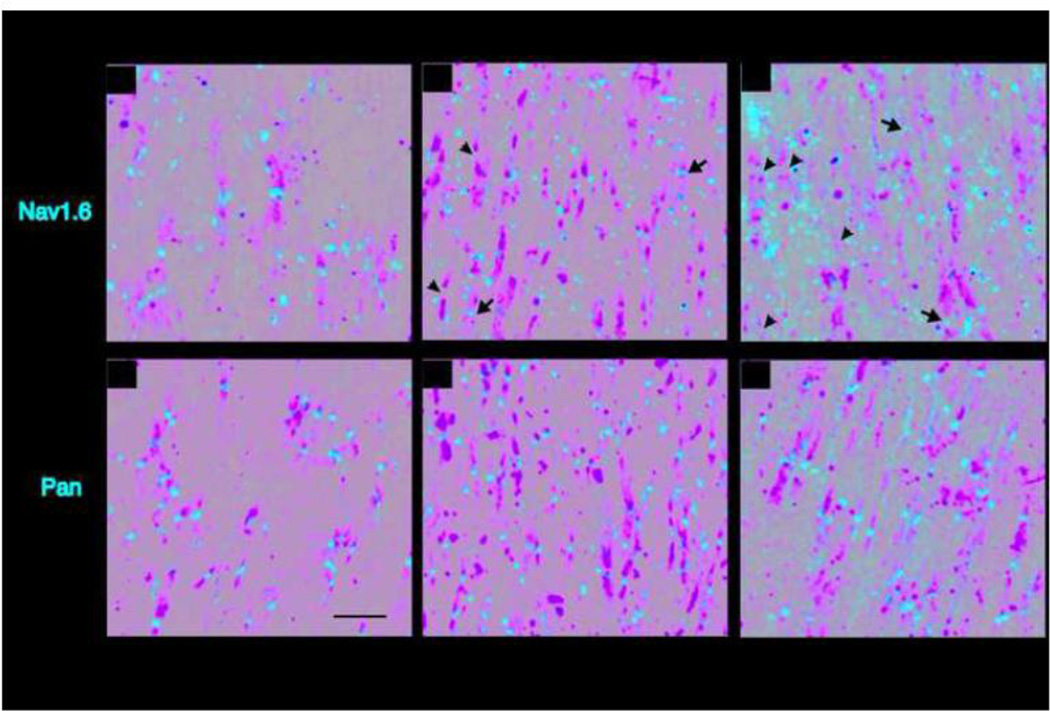
Figure 4. Nav1.6Δ1750 is not detectable at the axon initial segment (AIS).
Axon initial segments of the layer II/III of the visual cortex from 4 month old mice were immunolabeled using anti-Ankyrin G (AnkG. Red). NaV1.6 staining is localized to the distal AIS in wild-type mice (C), while AnkG-NaV1.6 co-localization was absent in ΔI/ΔI mice (F). A pan-NaV antibody (green) reveals sodium channels distributed along the length of the AIS in both WT and ΔI/ΔI. 8 micron optical sections are shown as sum intensity z-stack of 23 slices. Scale bar 10 microns.
Whole cell recordings from cerebellar Purkinje cells
Parasagittal cerebellar slices (300 micron) from 9 mutant and 5 littermate control mice at 5 weeks of age were prepared by vibratome sectioning in ice-cold solution as previously described (Shakkottai et al., 2011; Luna-Cancalon et al., 2014). Briefly, slices were incubated at 33°C in artificial CSF (ACSF) containing 125 mM NaCl, 3.5 mM KCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 2 mM CaCl2, 1 mM MgCl2 and 10 mM glucose, bubbled with 5% CO2/95% O2 for 45 min. Recordings from Purkinje neurons were visualized with infrared differential interference contrast (IR-DIC) optics on a Nikon upright microscope. Borosilicate glass patch pipettes (with resistances of 3–4 MΩ) were filled with internal recording solution containing: 119 mM K Gluconate, 2 mM Na Gluconate, 6 mM NaCl, 2 mM MgCl2, 0.9 mM EGTA, 10 mM HEPES, 14 mM Tris-Phosphocreatine, 4 mM MgATP, 0.3 mM Tris GTP. Whole-cell recordings were made in ACSF at room temperature 1–5 h after slice preparation using an Axopatch 200B amplifier, Digidata 1440A interface and pClamp-10 software (Molecular Devices) as previously described (Shakkottai et al., 2011; Luna-Cancalon et al., 2014).
Voltage data were acquired in the fast current clamp mode of the amplifier and filtered at 2 kHz. The fast current-clamp mode is necessary to reduce distortion of action potentials observed when patch-clamp amplifiers are used in current-clamp mode (Magistretti et al., 1996). We could obtain stable recordings without oscillations in fast current-clamp mode with electrode resistances above 3 megaohms, as has been previously reported (Swensen and Bean, 2003). The series resistance was 14.1 ± 1.2 mega ohms for wildtype cells (n=10) and 15.2 ± 1.0 for mutant cells (n=20); p=0.29 by the unpaired t-test assuming Gaussian distribution, with Welche's correction as we did not assume equal standard deviation. The capacitance was 581 ± 44 picofarads for wildtype cells (n=10) and 477 ± 31 for mutant cells (n=20); p=0.07, statistics as in previous sentence. Cells were held at −90 mV and injected with 1 second depolarizing current steps of 50 pA. Data were digitized at 10 kHz. Voltage traces were corrected for a 10 mV liquid junction potential.
Channel activity in transfected cells
Nucleotides c.5248–5250 were deleted from the tetrodotoxin (TTX)-resistant derivative of the murine NaV1.6 cDNA clone NaV1.6R by site directed mutagenesis as previously described (Tyrrell et al., 2001; Lampert et al., 2008; Yang et al., 2013). The neuron-derived cell line ND7/23 was co-transfected with the Nav1.6 cDNA and an eGFP cDNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as previously described (Sharkey et al., 2009). Twenty four hours after transfection, ND7/23 cells with robust green fluorescence signal were selected for patch-clamp. Whole-cell voltage-clamp recordings were performed at room temperature (20–22°C) using an Axopatch 200B amplifier (Axon Instruments, Foster City, CA). Cells were held at −120 mV, and a series of 100 ms step depolarizations were applied to activate sodium channels.
Nerve Conduction Velocity was recorded as previously described (Lenk et al., 2011). Five heterozygous Scn8aΔ1750/+ and 5 wildtype littermates were anesthetized with isoflurane and placed under a heating lamp to maintain body temperature at 32°C. Recordings were obtained using a Nicolet VikingQuest portable system and Nicolet disposable EEG needles. Sural nerve conduction velocities were obtained by stimulating proximally over a 3 cm region. Motor nerve conduction velocity was obtained by stimulating sciatic nerve and recording from muscle to isolate the motor component of the sciatic nerve.
Grip Strength was measured using a grip strength meter (Chatillon E-DFE-002; Columbus, OH) with a metallic grid (12 × 10 cm) as described (Wagnon et al., 2015).
Open Field behavior was assayed as previously described (McKinney et al., 2008). The open field chamber was a white acrylic box (75 × 75 x 30 cm) in a room lit by indirect white light. Wildtype (n = 8) and heterozygous mutant mice (n = 11) were placed singly in the center of the chamber and allowed to explore for 5 minutes. The open field was divided into a center zone and a peripheral zone by dividing the chamber into 64 squares of equal size, with a peripheral zone one square wide around the periphery of the chamber, and the remaining area comprising the center zone. Distance and time traveled in the center and periphery were calculated.
Immunostaining of axon initial segments (AIS)
Freshly dissected brains were cut in half sagittally, immersion fixed in 1% paraformaldehyde in PBS for 2 hours followed by immersion in 40% sucrose in PBS at 4°C. 20 µm sagittal cryosections were cut, with control and experimental genotypes collected onto the same slide. Sections were permeabilized with 0.4% triton X-100 diluted in tris buffered saline (TBS. 25 mM Tris, 140 mM NaCl, 3 mM KCl, pH 7.5) for 15 minutes, followed by blocking with 10% normal goat serum (NGS), 1% bovine serum albumin in TBS with 0.1% triton X-100 for 1 hour. Primary antibodies were diluted in TBS with 0.1% triton X-100 and 2% NGS and incubated at 4°C overnight. Primary antibodies were mouse IgG1 anti-NaV1.6 1:200 (Neuromab. Clone K87/A10), mouse IgG1 anti-pan sodium channel 1:200 (Sigma. Clone K58/35) and rabbit anti-AnkG 1:150 (Santa Cruz sc-28561). After washing in TBS and 10 minutes in blocking solution, secondary antibodies, diluted 1:500 in TBS with 1% NGS, were incubated for 1 hour. Secondary antibodies were goat anti-mouse IgG1 Alexa Fluor 488 conjugate, and goat anti-rabbit Alexa Fluor 594 conjugate (Life Technologies).
Immunostaining of nodes of Ranvier
Optic nerves were dissected, fixed in 4% paraformaldehyde for 30 min, cryoprotected in 15% and 30% sucrose and cryosectioned longitudinally at 10 microns. Sections were mounted on chrom-alum coated coverslips, dried, and permeabilized in 0.1M phosphate buffer containing 0.3% Triton X-100 and 10% goat serum (PBTGS). Primary antibodies were monoclonal anti-pan Na+ channels raised against a highly conserved epitope in the inactivation gate that labels all voltage-dependent mammalian Na+ channels (Rasband et al., 1999), and NeuroMab monoclonal anti-Nav1.6 clone K87A/10 (distributed by Antibodies, Inc., Davis, CA) that is specific for immunostaining of Nav1.6 (Sharkey et al., 2009; Osorio et al., 2010). Caspr was stained with a polyclonal antibody (Peles et al., 1997). Optic nerve sections were examined on an Olympus FV1000 laser scanning confocal microscope and images analyzed with Fluoview and Image-Pro.
Immunostaining of Neuromuscular Junctions was carried out as previously described (Joyce et al., 2015). Freshly dissected extensor digitorum longus (EDL) muscles were pinned in a gentle stretch, immersion fixed in 4% paraformaldehyde in PBS for 30 minutes, rinsed in PBS, cryoprotected in 40% sucrose in PBS at 4°C for > 24 hours, and cryosectioned longitudinally at 20 µm to generate a complete set of serial sections. Sections were blocked by incubation for 1 hour in TBS (0.25M Tris pH 7.5, 0.14 M NaCl, 3 mM KCl) containing 0.2% Triton X-100 (TBS+) and 3.5% M.O.M. (mouse on mouse) blocking reagent (Vector Labs). Sections were incubated for 30 min with a combination of two primary antibodies, anti-synaptic vesicle (SV2, Developmental Studies Hybridoma Bank) and anti-neurofilament, 165 kDa (2H3, Developmental Studies Hybridoma Bank), at 1:10 dilution in TBS+ with 7.5% M.O.M protein concentrate (Vector Labs). Sections were incubated for 10 min with 1:250 dilution of the secondary antibody, M.O.M. biotinylated anti-mouse (Vector Labs), diluted in TBS+ containing 7.5% M.O.M protein concentrate and then labeled for 30 min in TBS+ containing 1:200 dilution of streptavidin AlexaFluor 488 Conjugate (Invitrogen), 0.01 µg/ml α-Bungarotoxin-tetramethylrhodamine (α-BTX) (Sigma) and DAPI for nuclear staining. Sections were examined on an Olympus BX-51 fluorescence microscope. Muscle was prepared from two wildtype and two mutant animals.
RESULTS
Identification of a new mouse mutant with impaired movement and reduced muscle strength
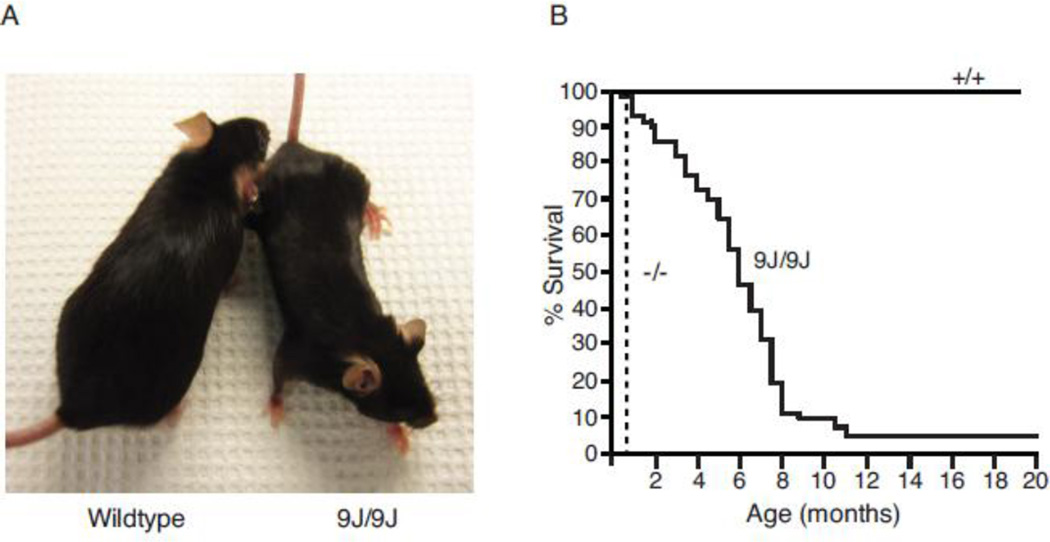
The spontaneous recessive mutation arose on strain BALB/cJ at the Jackson Laboratory in 2007. Heterozygous mutants, which are viable and fertile, were backcrossed to strain C57BL/6J for 10 generations. Homozygotes are born in the predicted Mendelian proportion but develop an early onset, progressive movement disorder. Tremor and ataxia begin between the second and third weeks of postnatal life and progress with age. Dystonic movements begin at 6 months of age and become more severe in the final months. The body weight of homozyous mutants is reduced to 70% that of wildtype littermates throughout the lifespan (Figure 1A). Mutant mice remain ambulatory with an abnormal, lurching gait (Supplemental Video 1). Fifty % of homozygotes survive beyond 6 months but only 5% survive beyond 11 months (Figure 1B). Spontaneous convulsive seizures were not seen in heterozygous or homozygous mice.
Figure 1. A novel mutant mouse with reduced body size and shortened lifespan.
A) Reduced size of mutant compared with wildtype littermate at 8 months of age. B) Reduced longevity of Scn8a9J/9J mice. None of the wildtype littermates died during the period of observation. Survival of mutant mice up to 18 months of age has been observed. The dotted line represents the survival of Scn8a null mice.
Muscle weakness is evident and there is a 50% reduction of grip strength from 217+ 10 g (n=4) in unaffected littermates to 96 ± 18 g (n=3) in homozygous mutants. Clinical features of the new mutant thus include early onset tremor, ataxic gait, muscle weakness, progressive dystonia, impaired mobility and reduced lifespan.
Genetic noncomplementation demonstrates allelism with Scn8a
Female recipients of ovary transplants from affected mutants were crossed to males from strain CAST/Ei and the obligate heterozygous F1 offspring were intercrossed to generate F2 mice for genetic mapping. Ninety-four % of affected F2 mice (16/17) were homozygous for the non-CAST allele of markers D15Mit14 and D15Mit16 on Chromosome 15, indicating linkage of the mutated gene to Scn8a. Heterozygous mutants were test-crossed with Scn8amed/+ heterozygotes carrying a null allele of Scn8a (Kohrman et al., 1996a). Two of the 5 offspring exhibited tremor and ataxia, demonstrating non-complementation of Scn8amed by the new mutation, which was designated Scn8a9J (www.informatics.jax.org/allele/MGI:3838627).
Three base pair in-frame deletion in Scn8a
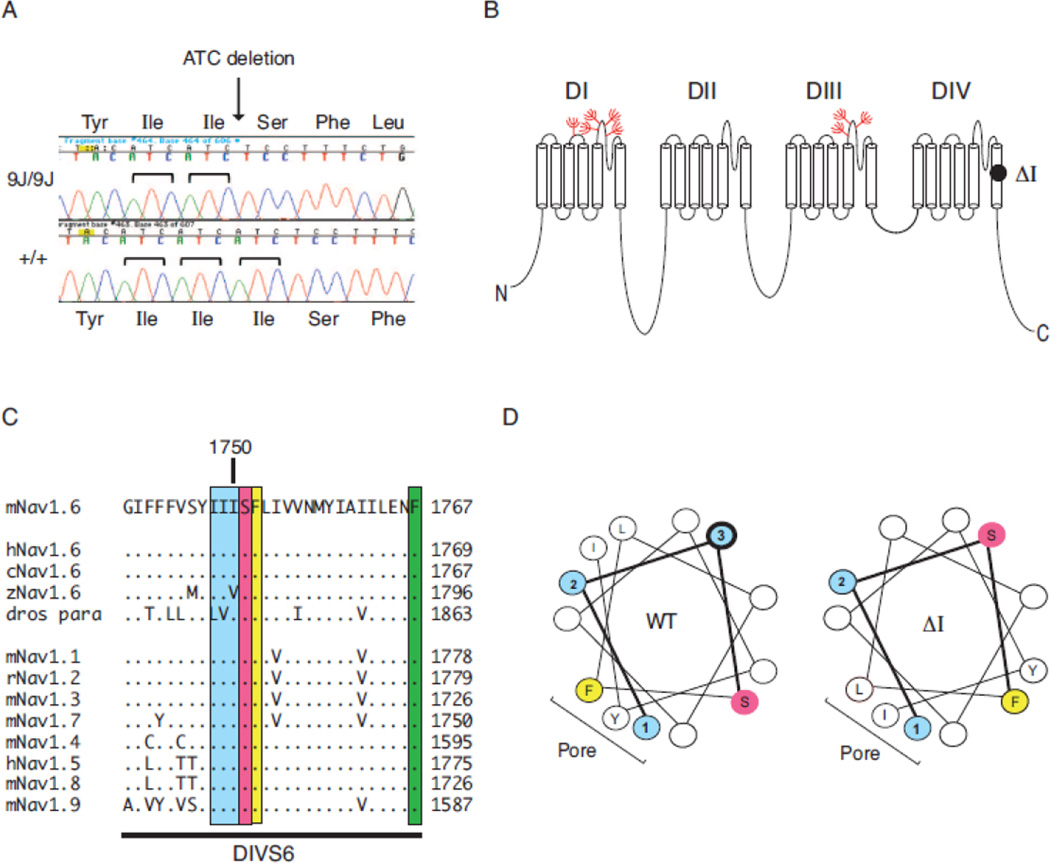
The 26 protein-coding exons and 2 alternatively spliced exons of Scn8a were amplified from homozygous mutant genomic DNA and subjected to Sanger sequencing. The 3 base pair deletion c.5248_5250delATC was identified in the final exon of the gene (Figure 2A). This in-frame deletion removes isoleucine codon 1750 in transmembrane segment DIVS6 (Figure 2B). This residue is located in an isoleucine3 repeat that is evolutionarily conserved in vertebrate Scn8a genes and paralogous gene family members (Figure 2C). Deletion of isoleucine 1750 alters the secondary structure of the distal half of the α-helix in transmembrane segment DIVS6, as shown in the helical wheel diagram (Figure 2D). The helical hydrophobic moment, a measure of amphiphilicity, is changed from 1.04 at 105° in the wildtype channel to 2.95 at 15.4° in the mutant channel (Supplement Figure 1, helical wheel projection program: www.rzlab.ucr.edu/script/wheel/wheel/cgi) (Zidovetzki et al., 2003).
Figure 2. Deletion of isoleucine codon 1750 of Scn8a.
A) Partial sequence chromatogram of exon 27 amplified from genomic DNA of homozygous mutant and wildtype control. The 3 bp deletion in the mutant removes one of the three adjacent ATC codons. B) Location of the deleted residue in transmembrane segment DIVS6 of Nav1.6. Location of glycosylation sites (red). C) The (Ile)3 repeat is evolutionarily conserved in vertebrate sodium channels. The Drosophila homolog contains conservative substitutions of leucine and valine for two isoleucine residues. Dots represent amino acid identity. D) Helical wheel representation of transmembrane segment DIVS6 beginning with isoleucine 1748. Blue, Isoleucine residues 1748–1750 (labeled 1–3). Yellow, phenylalanine 1752; pink, hydrophilic residues serine 1751 and asparagine 1757. The image was generated with the helical wheel projection program rzlab.ucr.edu/script/wheel/wheel/cgi.
The deletion of isoleucine 1750 is predicted to displace three critical residues of Nav1.6: Serine 1751, Phenylalanine 1752 and Phenylalanine 1769. Serine 1751 is located at the putative gating hinge involved in channel opening in the cardiac channel Nav1.5, in which it is designated serine 1759 (Figure 2, Supplement Figure 3) (Wang et al., 2005). The residue corresponding to Ser 1751 is Thr206 in the bacterial sodium channel NavMs, which is located at the start of the twist in S6 that is implicated in activation gate opening (Bagneris 2015); this residue corresponds to Ser1579 in Nav1.4 (Supplemental Figure 3). The residue corresponding to Nav1.6 phenylalanine 1752 is phenylalanine 1764 in Nav1.2, which is involved in fast inactivation (McPhee et al., 1994; McPhee et al., 1995; Oelstrom et al., 2014). Methionine 221 in bacterial channel NavAb seals the intracellular gate (Payandeh et al., 2011); this residue corresponds to phenylalanine 1769 in Nav1.6 (Supplemental Figure 3). Met222, the corresponding residue in bacterial channel NavMs, effectively corresponds to the location of the activation gate (Bagneris 2015). The Nav1.6Δl750 mutation thus disrupts the orientation of several known functional sites.
Incomplete glycosylation of Nav1.6Δ1750
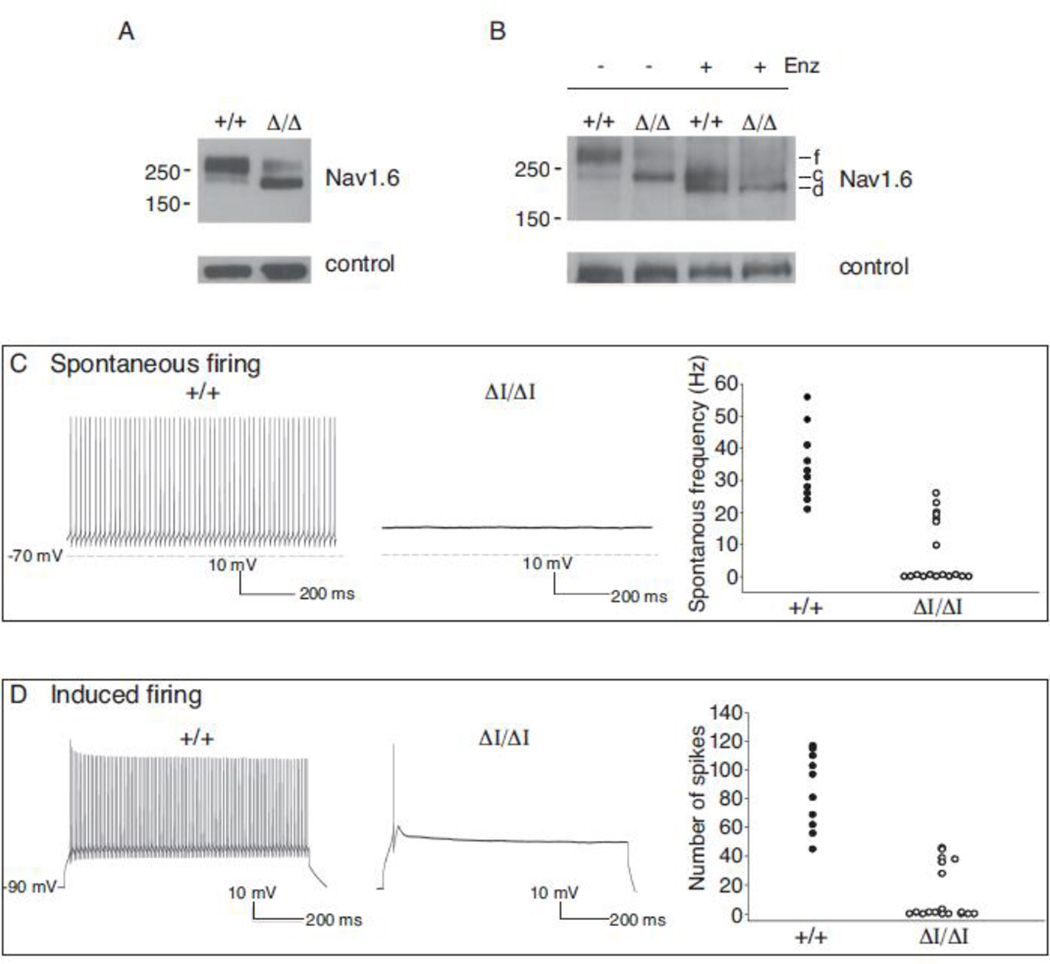
Glycosylation of the closely related sodium channel Nav1.2 (nonglycosylated MW 228 kDa) involves core glycosylation in the ER to an apparent MW of 240 kDa, followed by modification in the Golgi apparatus to a fully glycosylated MW of 260 kDa (Schmidt and Catterall, 1986). Nav1.6 has a nonglycosylated MW of 225 kDa and contains 8 predicted N-glycosylation sites (Figure 2B), but its glycosylation status has not been reported. Analysis of brain membrane preparations by Western blotting demonstrated that wildtype Nav1.6 migrates at 260 kDa, corresponding to the full glycosylation product of Nav1.2, while the Nav1.6Δ1750 protein migrates with an apparent MW of 240 kDA, corresponding to core glycosylation only (Figure 3A). Incubation of wildtype Nav1.6 for 1 hour with the N-glycosidase PNGaseF1 produced products with apparent MW of 240 kDa and 225 kDa (Figure 3B), corresponding to core-glycosylated and non-glycosylated Nav1.2. Incubation of Nav1.6Δ1750 with PNGaseF1 produced a product of 225 kDa, corresponding to conversion from core glycosylation to nonglycosylated protein (Figure 3A, right). The data indicate that glycosylation of Nav1.6Δ1750 is limited to core glycosylation (240 kDa).
Figure 3. Partial glycosylation of Nav1.6Δ1750 and reduced channel activity.
(A) Brain membrane proteins were fractionated on acrylamide gradient gels and immunostained with polyclonal anti-Nav1.6 (Alamone #ASC-009). The apparent molecular weight of the wildtype channel corresponds to the fully glycosylated protein (f). The mutant channel migrates with lower apparent molecular weight. Digestion with peptide N-glycosidase F converted the wildtype channel to two smaller products corresponding in mobility to core-glycosylated (c) and de-glycoslyated (d) proteins. f, fully glycosylated; c, core glycosylated; d, deglycosylated. Loading controls: left, α-tubulin; right, FIG 4. (B and C) Repetitive firing of Purkinje cells in cerebellar slices is impaired in Scn8a9J/9J mutant mice. The absence of repetitive firing by mutant cells is similar to previous observations of Nav1.6 null cells (Raman et al., 1997). (D) Current density in ND7/23 cells transfected with Nav1.6-Δ1750 cDNA or wildtype Nav1.6 cDNA, alone or together with FGF13b (F).
Impaired channel activity of Nav1.6Δ1750
To evaluate the impact of the Nav1.6Δ1750 on channel activity in neurons from mutant mice, we examined the firing pattern of cerebellar Purkinje neurons. Repetitive firing of Purkinje cells is dependent on Nav1.6, as indicated by the reduction in spontaneous and induced firing rates in cells prepared from Scn8a null mice (Raman et al., 1997; Khaliq et al., 2003; Sharkey et al., 2009). Purkinje cells from wildtype littermate controls exhibited an average spontaneous firing frequency of 36 ± 4 Hz (n=10 cells) (Figure 3C), and an induced firing rate after depolarizing current injection that exceeded 86 ± 8 spikes/sec (n=10) (Figure 3D), consistent with previous reports (Khaliq et al., 2003). In contrast, the majority of cells from Scn8a9J mice (11/18) did not exhibit any spontaneous firing (Figure 3C). After current injection, only 6/18 mutant cells fired, and those generated only one or a few spikes (Figure 3D). These observations demonstrate that the Nav1.6Δ1750 mutation results in substantial loss of in vivo channel activity, with an effect in Purkinje cells that is comparable in severity to the complete loss of Nav1.6 activity in Scn8a null cells (Raman et al., 1997).
To confirm the reduced sodium channel activity of the mutant, sodium currents were recorded after transfection of wildtype and mutant Nav1.6 cDNA into the neuron-derived cell line ND7/23. Cells expressing the wildtype cDNA exhibited robust sodium current density of 46 ± 17 pA/pF (n=4), but current density in cells transfected with Nav1.6Δ1750 (n=6) was below the threshold for detection. We attempted to enhance the activity of the mutant channel by reducing the temperature of cell culture from 37°C to 30°C and co-transfection of FGF13, a non-secreted member of the FGF superfamily that interacts with Nav1.6 (Wittmack et al., 2004). The activity of wildtype Nav1.6 was increased to 93 ± 25 pA/pF (n=3), but the activity of the mutant channel (n=8) remained below the level of detection. The mutant channel was also inactive in transfected HEK293 cells. Thus, the Nav1.6-Δ1750 mutation results in a severely hypomorphic channel with profound reduction in activity.
Localization to the axon initial segment of cortical neurons
Localization of Nav1.6 to the axon initial segment (AIS) is responsible for its role in initiation of action potentials, and is thought to be important for repetitive firing by Purkinje cells. To determine whether the mutant Nav1.6Δ1750 protein is correctly localized to the AIS, sections of visual cortex layer II/III were immunostained for Nav1.6 and Ankyrin G. In sections from wildtype brain, co-localization of Ankyrin G and Nav1.6 was evident (Figure 4, top), but in homozygous mutant mice, Nav1.6 staining was missing from the AIS. Even at 20 months of age, Nav1.6 was not detectable at the AIS (Supplemental Figure 4). To determine whether Nav1.6 is replaced by other voltage-gated sodium channels at the AIS, sections were stained with pan-sodium channel antibody that recognizes all neuronal sodium channels, including Nav1.1 and Nav1.2. Strong staining with the pan-antibody demonstrates that Nav1.6 is replaced at the AIS of mutant mice.
Nav1.6Δ1750 at CNS nodes of Ranvier
To assess the localization of the mutant channel to nodes of Ranvier, optic nerves were sectioned longitudinally and immunostained for Nav1.6. Nodes were defined by the presence of two paranodes immunostained for the paranodal protein Caspr. In wildtype nerves at 7 months of age, 97% of nodes (207/213) were positive for Nav1.6 and Nav1.6 staining filled the gap between paranodes (Figure 5A). In 7 month old homozygous mutant mice, only 60% of nodes (68/113) were labeled with anti-Nav1.6 (Fig. 5B, arrows) with the remaining 40% of nodes lacking Nav1.6 immunoreactivity (arrowheads). Nav1.6 staining did not completely fill the nodal gap in one third of the labeled nodes from mutant nerves. To determine whether Nav1.6 accumulates at nodes with increasing age, we examined optic nerve from 10 month old mice. We observed exacerbation of the defect, with immunolabeling of only 38% (29/77) of mutant nodes, with only 4% exhibiting normal labeling (Figure 5C). In addition, the 10 month old nerves contain many extra-nodal concentrations of Nav1.6 that appear to result from delocalization or degeneration of the mutant channel.
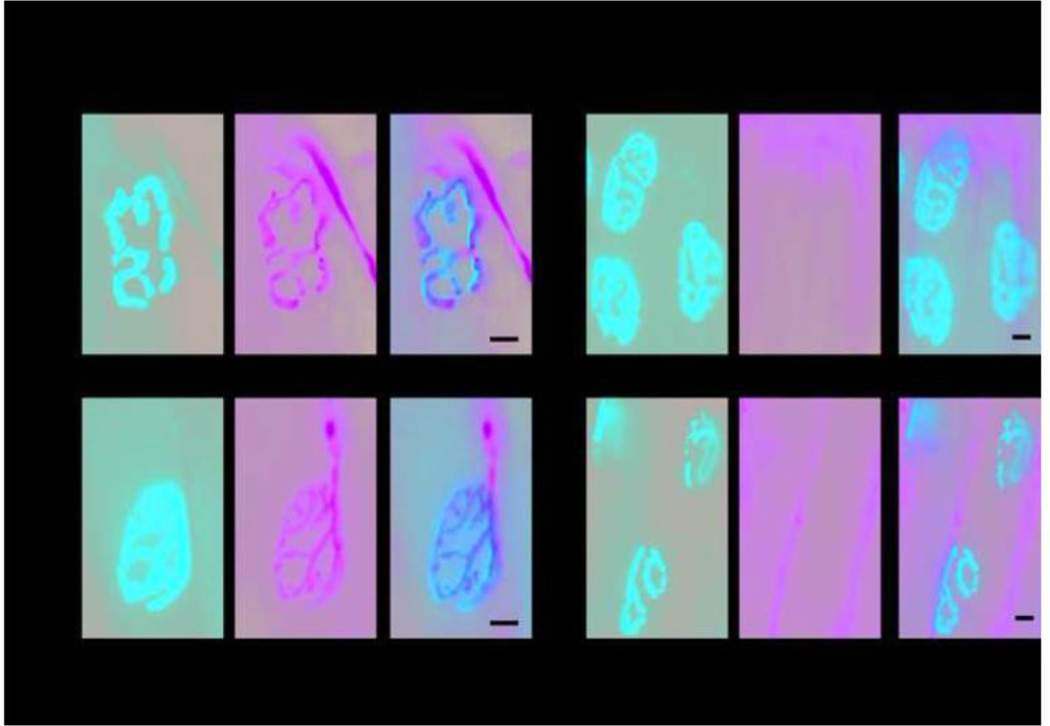
Figure 5. Localization of Nav1.6Δ1750 at nodes of Ranvier in the optic nerve.
Longitudinal cryosections of optic nerve were immunostained with anti-Caspr to identify the paranodes (green), and with specific antiserum to Nav1.6 (red, top panels), or nonspecific pan Na+ channel antiserum that detects all mammalian sodium channels (red, bottom panels). Arrows mark nodes containing Nav1.6. Arrowheads mark empty nodes. Scale bar, 5 microns.
To determine whether Nav1.6 is replaced by other sodium channels in the mutant nodes, we stained optic nerves with a monoclonal antibody that recognizes all mammalian voltage-dependent sodium channels ("pan" antibody) (Rasband et al., 1999). Essentially all nodes in mutant and wildtype optic nerve were labeled with the pan-sodium channel antibody (Figure 5D–F).
These data indicate that nodes of Ranvier in Scn8adel1750 homozygotes are partially deficient in Nav1.6, and that other voltage-gated sodium channels replace Nav1.6 at the deficient nodes. The "pan" antibody does not distinguish between Nav1.1, Nav1.2 and Nav1.3, but it was previously demonstrated that both Nav1.1 and Nav1.2 can localize at nodes of Ranvier in mice deficient in Nav1.6 (Kearney et al., 2002) (Van Wart and Matthews, 2006).
Nerve conduction velocity and open field behavior
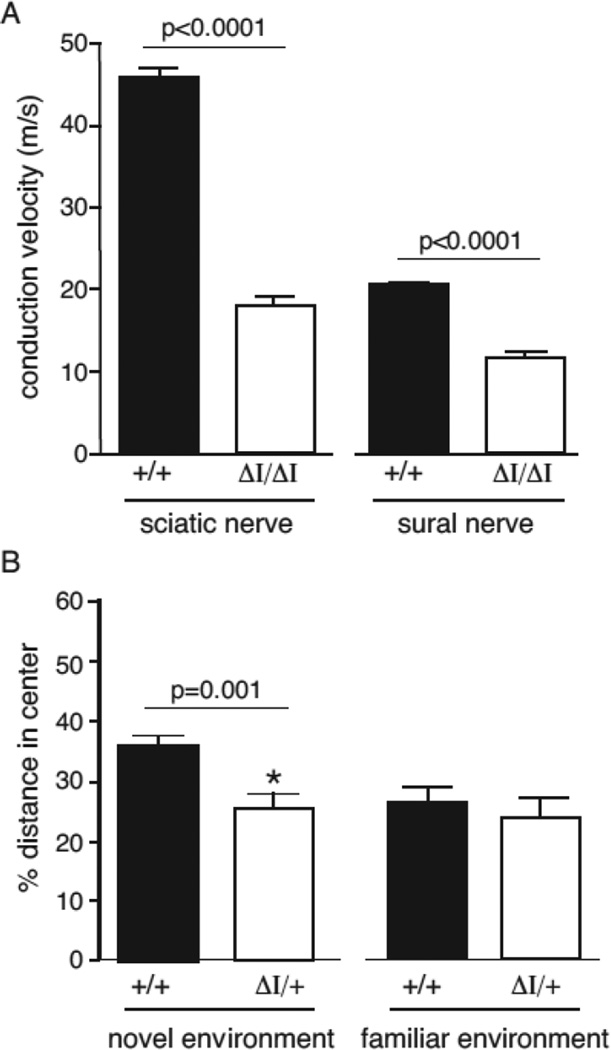
To assess nerve function in the mutant mice, we measured conduction velocity in two peripheral nerves. The motor component of the sciatic-tibial nerve was evaluated by stimulation of the nerve and recording from muscle. Compound action potentials were readily recorded, but conduction velocity was reduced by 50% in mutant nerve (Figure 6A). In sural nerve, a peripheral sensory nerve, conduction velocity was also reduced by 50% (Figure 4A).
Figure 6. Reduced nerve conduction velocity and impaired open field behavior.
(A) Nerve conduction velocity of sciatic and sural nerve from wildtype and homozygous mutant mice. Values are mean ± SEM (n=6). (B) Behavior of heterozygous Scn8a9J/+ mice (A/+) and wildtype mice in the open field test. Left, heterozygous mice avoid the center of the open field, indicative of elevated anxiety (p= 0.001, unpaired t-test). Right, in a familiar environment, the behavior of heterozygous mice does not differ from wildtype littermate controls. The total distance traveled did not differ, but the distance traveled by heterozygous mutants in the center of the field was lower in the novel environment (Suppl. Figure 2.)
Heterozygous Scn8a null mice exhibit impaired performance in the open field test (McKinney et al., 2008). We therefore examined the behavior of Nav1.6Δ1750 heterozygotes in this behavioral test. Performance of the heterozygotes in a familiar environment did not differ from wildtype littermates (Figure 6B). However, in an unfamiliar environment the distance traveled by Scn8a9J heterozygotes was significantly reduced (p = 0.001, unpaired t-test) (Figure 6B, Supplement Figure 2). This behavioral abnormality reproduces that of heterozygous null mice, supporting the evidence above that Nav1.6Δ1750 lacks channel activity.
Structure of the neuromuscular junction (NMJ)
It was recently reported that the neuronal and muscular components of the NMJ are defective in Scn8a−/− null mice (Caillol et al., 2012). The null NMJs lack the characteristic “pretzel” shape of the mature neuromuscular junction and the nerve terminals fail to develop complex arborization. To determine whether defects of the NMJ contribute to pathogenesis of Scn8a9J/9J mice, we carried out quantitative analysis of the NMJs at 1 year of age, when the mice exhibit compromised motor function and are near the end of their lifespan (Figure 1B). NMJs were quantitated after immunostaining of a complete set of serial sections of the extensor digitalis longus muscle of the hind limb. The total number of NMJs was 980 in the mutant muscle and 986 in the wildtype muscle. The proportion of normally innervated NMJs was 58% in mutant muscle and 55% in wildtype muscle, and their morphology did not differ between mutant and wildtype (Figure 7A). The proportion of denervated NMJs was 37% in the mutant muscle and 36% in wildtype muscle (Figure 7B), with the remainder partially innervated. The NMJs of the diaphragm muscle also exhibited normal number and morphology (data not shown). Thus defects in the NMJ do not appear to contribute to the movement disorder or reduced lifespan of Scn8a9J/9J mice.
Figure 7. Morphology of the neuromuscular junction.
Serial sections of the extensor digitalis longus muscle of the hind limb from 1 year old Scn8a9J/9J mice and wild-type littermates were immunostained with antibodies to the neuronal proteins SV2 and 2H3 and with α-bungarotoxin to detect the acetylcholine receptor in the muscle endplate. The total number, morphology, and proportion of innervated NMJs did not differ between mutant and wildtype mice. A. Normal morphology of innervated neuromuscular junctions demonstrating the complex perforated morphology and close apposition of pre and post-terminal components. B. Regions with denervated endplates were apparent in mutant and wildtype mice. Scale bar, 10 µm.
DISCUSSION
Scn8a9J is a hypomorphic allele with a novel recessive phenotype
The properties of mice carrying the new allele of Scn8a described here differ from previously described mutants (O'Brien and Meisler, 2013). Scn8a9J homozygotes are viable, unlike null mutants lacking Nav1.6 protein, and exhibit chronic tremor and a distinctive lurching gait with subsequent development of progressive dystonia. The channel activity of the mutant Nav1.6 protein is greatly reduced in comparison with wildtype channel. The loss of spontaneous and induced repetitive firing by cerebellar Purkinje cells mimicks the cells from Scn8a null mutants (Raman et al., 1997; Khaliq et al., 2003). Nerve conduction velocity and open field behavior of heterozygous Scn8a9J/+ mice also recapitulate the null heterozygote (Kearney et al., 2002; McKinney et al., 2008). Nonetheless, the clinical phenotype is much less severe than previously characterized null mutants that lack channel protein as a consequence of protein truncations, impaired trafficking, or splice site mutations (Burgess et al., 1995; Kohrman et al., 1996b; Kearney et al., 2002; Sharkey et al., 2009).
Scn8a9J is phenotypically less severe than the hypomorphic Scn8amedJ mutant
In the Scn8amedJ mouse, a splice site mutation reduces the level of Nav1.6 protein to 10% of wildtype level, with retention of wildtype amino acid sequence and activity (Kohrman et al., 1996a; Kearney et al., 2002). The level of sodium channel activity in a heterologous system is similar for the Nav1.6Δ1750 mutant protein. Nonetheless, the clinical disorder in Scn8amedJ mice is much more severe than for Scn8a9J mice. For example, Scn8amedJ mice cannot walk or support their body weight, while Scn8a9J mice are ambulatory (see comparison in Supplemental video). The dystonia in Scn8amedJ mice exhibits earlier onset and greater severity (Sprunger et al., 1999). One difference that may contribute to the more robust phenotype of Scn8a9J mice is their higher abundance of Nav1.6 protein.
Effects of the Scn8a9J mutation on protein structure
Deletion of residue 1750 alters the orientation of several residues with known functions in channel activation and inactivation, as discussed in Results. The orientation of 17 downstream amino acids within the DIVS6 α-helix is altered, and the identity of residues facing the channel pore changes from Ile1748-Phe1752-Tyr1759 to Ile1748-Leu1752-Ile1759 (Figure 2D, Supplement Figure 1). The strength and orientation of the hydrophobic moment are also changed.
Phe1752 is a target for binding of high affinity voltage-dependent sodium channel inhibitors (Mike and Lukacs, 2010). Drug binding to Phe1752 inhibits movement of the gating charges and thereby disrupts channel function (Muroi and Chanda, 2009). Phe1752 also plays a role in channel fast inactivation (McPhee et al., 1995) and in interaction with the sodium channel β4 subunit (Wang et al., 2006). The substitution of a leucine side chain for Phe1752 is thus predicted to have major consequences for channel function.
Ser1751 (Figure 2B, Supplement Figure 3) is thought to be the site around which the DIVS6 α-helix twists during channel opening (Oelstrom et al., 2014; Bagneris et al., 2015). By substituting a phenylalanine residue at this position, the Δ1750 mutation could impair the productive splaying of the S6 transmembrane segment, resulting in abortive opening of the activation gate. This view is consistent with the observed loss of channel activity. Ser1751 has also been identified as a potential gating-hinge during slow inactivation (Wang et al., 2005).
Phe1769 is predicted by the crystal structure to be the last residue of DIVS6 and to function as part of the activation gate (Payandeh et al., 2011; Bagneris et al., 2015). Modeling of Nav1.7 also indicates that this residue contributes to the activation gate (Lampert et al., 2008; Yang et al., 2013). The Δ1750 mutation would move this residue into the membrane, substantially altering the geometry of the activation gate of Nav1.6.
In view of the critical roles of these residues in channel activation and inactivation, the impaired channel activity of Nav1.6Δ1750 in Purkinje cells is not surprising. A similar deletion in DIIS6 of SCN9A was shown to disrupt radial tuning of the domain II S6 helix, with helical displacement of an activation gate residue that pathologically altered activation (Yang et al., 2013).
Channel glycoslylation
The extensive glycosylation and sialylation of voltage gated ion channels is known to modulate channel activity (Tyrrell et al., 2001; Ednie and Bennett, 2012; Baycin-Hizal et al., 2014; Scott and Panin, 2014). Core-glycosylation of Nav1.5 and Nav1.7 does not prevent transport to the cell membrane (Laedermann et al., 2013; Mercier et al., 2015), but core glycosylated Nav1.5 appears to be inactive (Mercier et al., 2015). We found that the predominant form of Nav1.6Δ1750 is core glycosylated, with an apparent MW of 240 kDa. Reduced glycosylation is accompanied by loss of channel activity, consistent with the previous work on Nav1.5.
Subcellular localization of Nav1.6
A striking consequence of the Δ1750 mutation is the absence of detectable Nav1.6 from the AIS of cortical neurons in adult mice. It is not clear whether the defect is in trafficking or stabilization of the mutant channel at the AIS. Nav1.6 is replaced by other voltage-gated sodium channels at the mutant AIS. Since Nav1.6 is normally the major channel at the AIS of cerebellar Purkinje (Lorincz and Nusser, 2008), this impaired localization may contribute to the lack of repetitive firing observed in mutant Purkinje cells.
The concentration of Nav1.6 at nodes of Ranvier is reduced, with replacement by voltage-gated sodium channels. This is consistent with previous studies of nodal sodium channels. Nav1.2 is present in immature nodes of Ranvier but is replaced by Nav1.6 during nodal maturation (Boiko et al., 2001). In the Nav1.6 null mouse, both Nav1.1 and Nav1.2 can be detected at the axon initial segment and nodes of Ranvier (Van Wart and Matthews, 2006). Persistence of Nav1.2 at nodes of Ranvier was also observed in the hypomorphic Scn8amedJ mutant (Kearney et al., 2002).
Pathogenic model
The effects of the deletion of isoleucine 1750 in Nav1.6 include alteration of the positions of key residues within domain IV segment 6, reduction of channel activity, impaired glycosylation, loss of Nav1.6 protein at the axon initial segment and reduced Nav1.6 in nodes of Ranvier. These molecular deficits result in tremor, ataxia and dystonia in homozygous Scn8a9J/9J mice. De novo gain-of-function mutations of human SCN8A are responsible for epileptic encephalopathy (OMIM #614558) (Wagnon and Meisler, 2015). This work suggests SCN8A should also be considered as a candidate gene for recessively inherited movement disorders.
Supplementary Material
Highlights.
Spontaneous mutation of Scn8a generates a recessive movement disorder.
Deletion of residue 1750 alters the identity of pore-lining residues.
Sodium channel activity is reduced in Purkinje cells and ND7/23 cells.
Defective glycosylation and localization of Nav1.6 to nodes of Ranvier.
Mutant protein Nav1.6 is not detectable at the axon initial segment.
Acknowledgments
Supported by NIH grants R01 NS034509 (MHM), K08 NS072158 (VGS), R01 NS085054 (VGS) and P40 OD010972 to the Mouse Mutant Resource at the Jackson Laboratory. We thank Margaret Youngman for excellent technical assistance and Wan Tang for help with statistical analysis. Stephen Waxman and David Bergstrom provided helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bagneris C, et al. Structural model of the open-closed-inactivated cycle of prokaryotic voltage-gated sodium channels. J Gen Physiol. 2015;145:5–16. doi: 10.1085/jgp.201411242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baycin-Hizal D, et al. Physiologic and pathophysiologic consequences of altered sialylation and glycosylation on ion channel function. Biochem Biophys Res Commun. 2014;453:243–253. doi: 10.1016/j.bbrc.2014.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard MG, et al. De novo gain-of-function and loss-of-function mutations of SCN8A in patients with intellectual disabilities and epilepsy. J Med Genet. 2015;52:330–337. doi: 10.1136/jmedgenet-2014-102813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiko T, et al. Compact myelin dictates the differential targeting of two sodium channel isoforms in the same axon. Neuron. 2001;30:91–104. doi: 10.1016/s0896-6273(01)00265-3. [DOI] [PubMed] [Google Scholar]
- Burgess DL, et al. Mutation of a new sodium channel gene, Scn8a, in the mouse mutant ‧motor endplate disease'. Nat Genet. 1995;10:461–465. doi: 10.1038/ng0895-461. [DOI] [PubMed] [Google Scholar]
- Caillol G, et al. Motor endplate disease affects neuromuscular junction maturation. Eur J Neurosci. 2012;36:2400–2408. doi: 10.1111/j.1460-9568.2012.08164.x. [DOI] [PubMed] [Google Scholar]
- Caldwell JH, et al. Sodium channel Na(v)1.6 is localized at nodes of ranvier, dendrites, and synapses. Proc Natl Acad Sci U S A. 2000;97:5616–5620. doi: 10.1073/pnas.090034797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvill GL, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet. 2013;45:825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA, et al. Voltage-gated ion channels and gating modifier toxins. Toxicon. 2007;49:124–141. doi: 10.1016/j.toxicon.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Chen Y, et al. Functional properties and differential neuromodulation of Na(v)1.6 channels. Mol Cell Neurosci. 2008;38:607–615. doi: 10.1016/j.mcn.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JS, et al. Resurgent Na+ current: a new avenue to neuronal excitability control. Life Sci. 2011;89:564–569. doi: 10.1016/j.lfs.2011.05.016. [DOI] [PubMed] [Google Scholar]
- de Kovel CG, et al. Characterization of a de novo SCN8A mutation in a patient with epileptic encephalopathy. Epilepsy Res. 2014;108:1511–1518. doi: 10.1016/j.eplepsyres.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen LW. Hereditary motor end-plate disease in the mouse: light and electron microscopic studies. J Neurol Neurosurg Psychiatry. 1970;33:238–250. doi: 10.1136/jnnp.33.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ednie AR, Bennett ES. Modulation of voltage-gated ion channels by sialylation. Compr Physiol. 2012;2:1269–1301. doi: 10.1002/cphy.c110044. [DOI] [PubMed] [Google Scholar]
- Estacion M, et al. A novel de novo mutation of SCN8A (Nav1.6) with enhanced channel activation in a child with epileptic encephalopathy. Neurobiol Dis. 2014;69:117–123. doi: 10.1016/j.nbd.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce PI, et al. A novel SOD1-ALS mutation separates central and peripheral effects of mutant SOD1 toxicity. Hum Mol Genet. 2015;24:1883–1897. doi: 10.1093/hmg/ddu605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney JA, et al. Molecular and pathological effects of a modifier gene on deficiency of the sodium channel Scn8a (Na(v)1.6) Hum Mol Genet. 2002;11:2765–2775. doi: 10.1093/hmg/11.22.2765. [DOI] [PubMed] [Google Scholar]
- Khaliq ZM, et al. The contribution of resurgent sodium current to high-frequency firing in Purkinje neurons: an experimental and modeling study. J Neurosci. 2003;23:4899–4912. doi: 10.1523/JNEUROSCI.23-12-04899.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrman DC, et al. Mutation detection in the med and medJ alleles of the sodium channel Scn8a. Unusual splicing due to a minor class AT-AC intron. J Biol Chem. 1996a;271:17576–17581. doi: 10.1074/jbc.271.29.17576. [DOI] [PubMed] [Google Scholar]
- Kohrman DC, et al. A missense mutation in the sodium channel Scn8a is responsible for cerebellar ataxia in the mouse mutant jolting. J Neurosci. 1996b;16:5993–5999. doi: 10.1523/JNEUROSCI.16-19-05993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laedermann CJ, et al. beta1- and beta3-voltage-gated sodium channel subunits modulate cell surface expression and glycosylation of Nav1.7 in HEK293 cells. Front Cell Neurosci. 2013;7:137. doi: 10.3389/fncel.2013.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert A, et al. A pore-blocking hydrophobic motif at the cytoplasmic aperture of the closed-state Nav1.7 channel is disrupted by the erythromelalgia-associated F1449V mutation. J Biol Chem. 2008;283:24118–24127. doi: 10.1074/jbc.M802900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J, et al. The phenotypic spectrum of SCN8A encephalopathy. Neurology. 2015;84:480–489. doi: 10.1212/WNL.0000000000001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazniewska J, Weiss N. The "sweet" side of ion channels. Rev Physiol Biochem Pharmacol. 2014;167:67–114. doi: 10.1007/112_2014_20. [DOI] [PubMed] [Google Scholar]
- Lenk GM, et al. Pathogenic mechanism of the FIG 4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet. 2011;7:e1002104. doi: 10.1371/journal.pgen.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz A, Nusser Z. Cell-type-dependent molecular composition of the axon initial segment. J Neurosci. 2008;28:14329–14340. doi: 10.1523/JNEUROSCI.4833-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz A, Nusser Z. Molecular identity of dendritic voltage-gated sodium channels. Science. 2010;328:906–909. doi: 10.1126/science.1187958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Cancalon K, et al. Alterations in cerebellar physiology are associated with a stiff-legged gait in Atcay mice. Neurobiol Dis. 2014;67C:140–148. doi: 10.1016/j.nbd.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistretti J, et al. Action potentials recorded with patch-clamp amplifiers: are they genuine? Trends Neurosci. 1996;19:530–534. doi: 10.1016/s0166-2236(96)40004-2. [DOI] [PubMed] [Google Scholar]
- McKinney BC, et al. Exaggerated emotional behavior in mice heterozygous null for the sodium channel Scn8a (Nav1.6) Genes Brain Behav. 2008;7:629–638. doi: 10.1111/j.1601-183X.2008.00399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee JC, et al. A mutation in segment IVS6 disrupts fast inactivation of sodium channels. Proc Natl Acad Sci U S A. 1994;91:12346–12350. doi: 10.1073/pnas.91.25.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee JC, et al. A critical role for transmembrane segment IVS6 of the sodium channel alpha subunit in fast inactivation. J Biol Chem. 1995;270:12025–12034. doi: 10.1074/jbc.270.20.12025. [DOI] [PubMed] [Google Scholar]
- Mercier A, et al. Na1.5 channels can reach the plasma membrane through distinct N-glycosylation states. Biochim Biophys Acta. 2015;1850:1215–1223. doi: 10.1016/j.bbagen.2015.02.009. [DOI] [PubMed] [Google Scholar]
- Mike A, Lukacs P. The enigmatic drug binding site for sodium channel inhibitors. Curr Mol Pharmacol. 2010;3:129–144. doi: 10.2174/1874467211003030129. [DOI] [PubMed] [Google Scholar]
- Muroi Y, Chanda B. Local anesthetics disrupt energetic coupling between the voltage-sensing segments of a sodium channel. J Gen Physiol. 2009;133:1–15. doi: 10.1085/jgp.200810103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien JE, Meisler MH. Sodium channel (Na1.6): properties and mutations in epileptic encephalopathy and intellectual disability. Front Genet. 2013;4:213. doi: 10.3389/fgene.2013.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien JE, et al. Interaction of voltage-gated sodium channel Nav1.6 (SCN8A) with microtubule-associated protein Map1b. J Biol Chem. 2012;287:18459–18466. doi: 10.1074/jbc.M111.336024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelstrom K, et al. Evolutionarily conserved intracellular gate of voltage-dependent sodium channels. Nat Commun. 2014;5:3420. doi: 10.1038/ncomms4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio N, et al. Persistent Nav1.6 current at axon initial segments tunes spike timing of cerebellar granule cells. J Physiol. 2010;588:651–670. doi: 10.1113/jphysiol.2010.183798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J, et al. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peles E, et al. Identification of a novel contactin-associated transmembrane receptor with multiple domains implicated in protein-protein interactions. EMBO J. 1997;16:978–988. doi: 10.1093/emboj/16.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman IM, et al. Altered subthreshold sodium currents and disrupted firing patterns in Purkinje neurons of Scn8a mutant mice. Neuron. 1997;19:881–891. doi: 10.1016/s0896-6273(00)80969-1. [DOI] [PubMed] [Google Scholar]
- Rasband MN, et al. Dependence of nodal sodium channel clustering on paranodal axoglial contact in the developing CNS. J Neurosci. 1999;19:7516–7528. doi: 10.1523/JNEUROSCI.19-17-07516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royeck M, et al. Role of axonal NaV1.6 sodium channels in action potential initiation of CA1 pyramidal neurons. J Neurophysiol. 2008;100:2361–2380. doi: 10.1152/jn.90332.2008. [DOI] [PubMed] [Google Scholar]
- Rush AM, et al. Electrophysiological properties of two axonal sodium channels, Nav1.2 and Nav1.6, expressed in mouse spinal sensory neurones. J Physiol. 2005;564:803–815. doi: 10.1113/jphysiol.2005.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JW, Catterall WA. Biosynthesis and processing of the alpha subunit of the voltage-sensitive sodium channel in rat brain neurons. Cell. 1986;46:437–444. doi: 10.1016/0092-8674(86)90664-1. [DOI] [PubMed] [Google Scholar]
- Scott H, Panin VM. The role of protein N-glycosylation in neural transmission. Glycobiology. 2014;24:407–417. doi: 10.1093/glycob/cwu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakkottai VG, et al. Early changes in cerebellar physiology accompany motor dysfunction in the polyglutamine disease spinocerebellar ataxia type 3. J Neurosci. 2011;31:13002–13014. doi: 10.1523/JNEUROSCI.2789-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey LM, et al. The ataxia3 mutation in the N-terminal cytoplasmic domain of sodium channel Na(v)1.6 disrupts intracellular trafficking. J Neurosci. 2009;29:2733–2741. doi: 10.1523/JNEUROSCI.6026-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampanato J, et al. Functional effects of two voltage-gated sodium channel mutations that cause generalized epilepsy with febrile seizures plus type 2. J Neurosci. 2001;21:7481–7490. doi: 10.1523/JNEUROSCI.21-19-07481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprunger LK, et al. Dystonia associated with mutation of the neuronal sodium channel Scn8a and identification of the modifier locus Scnm1 on mouse chromosome 3. Hum Mol Genet. 1999;8:471–479. doi: 10.1093/hmg/8.3.471. [DOI] [PubMed] [Google Scholar]
- Swensen AM, Bean BP. Ionic mechanisms of burst firing in dissociated Purkinje neurons. J Neurosci. 2003;23:9650–9663. doi: 10.1523/JNEUROSCI.23-29-09650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell L, et al. Glycosylation alters steady-state inactivation of sodium channel Nav1.9/NaN in dorsal root ganglion neurons and is developmentally regulated. J Neurosci. 2001;21:9629–9637. doi: 10.1523/JNEUROSCI.21-24-09629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wart A, Matthews G. Impaired firing and cell-specific compensation in neurons lacking nav1.6 sodium channels. J Neurosci. 2006;26:7172–7180. doi: 10.1523/JNEUROSCI.1101-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramah KR, et al. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet. 2012;90:502–510. doi: 10.1016/j.ajhg.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, et al. Convulsive seizures and SUDEP in a mouse model of SCN8A epileptic encephalopathy. Hum Mol Genet. 2015;24:506–515. doi: 10.1093/hmg/ddu470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Meisler MH. Recurrent and Non-Recurrent Mutations of SCN8A in Epileptic Encephalopathy. Front Neurol. 2015;6:104. doi: 10.3389/fneur.2015.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, et al. Pathogenic mechanisms of recurrent epileptogenic mutations of SCN8A in epileptic encephalopathy. Annals of Clinical and Translational Neurology. 2015 doi: 10.1002/acn3.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GK, et al. Time-dependent block and resurgent tail currents induced by mouse beta4(154–167) peptide in cardiac Na+ channels. J Gen Physiol. 2006;127:277–289. doi: 10.1085/jgp.200509399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SY, et al. Tryptophan substitution of a putative D4S6 gating hinge alters slow inactivation in cardiac sodium channels. Biophys J. 2005;88:3991–3999. doi: 10.1529/biophysj.105.059352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmack EK, et al. Fibroblast growth factor homologous factor 2B: association with Nav1.6 and selective colocalization at nodes of Ranvier of dorsal root axons. J Neurosci. 2004;24:6765–6775. doi: 10.1523/JNEUROSCI.1628-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. Molecular architecture of a sodium channel S6 helix: radial tuning of the voltage-gated sodium channel 1.7 activation gate. J Biol Chem. 2013;288:13741–13747. doi: 10.1074/jbc.M113.462366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidovetzki R, et al. Transmembrane domains in the functions of Fc receptors. Biophys Chem. 2003;100:555–575. doi: 10.1016/s0301-4622(02)00306-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.