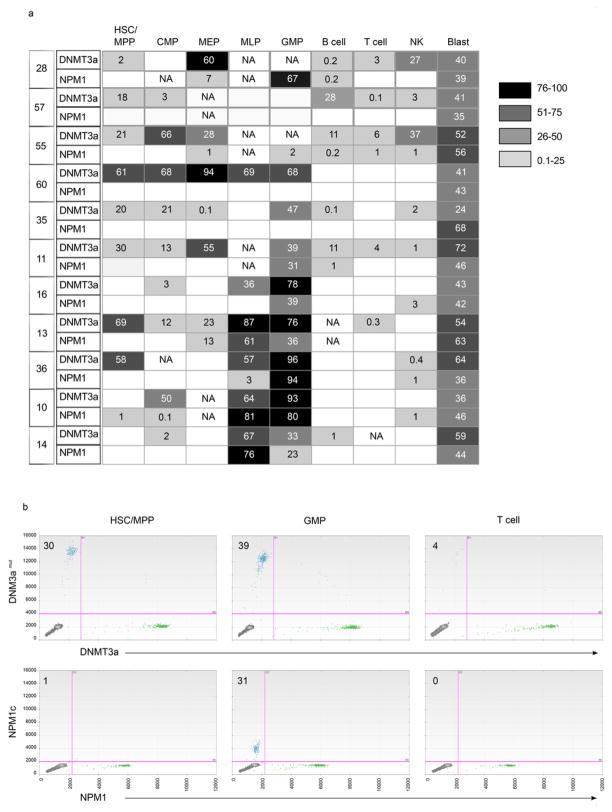
Extended Data Figure 2. Frequent occurrence of DNMT3A mutation without NPM1 mutation in stem/progenitor and mature lymphoid cells in AML patients at diagnosis.
a, Summary of the allele frequency (%) of DNMT3A and NPM1 mutations in stem/progenitor, mature lymphoid, and blast (CD45dim CD33+) cell populations from 11 AML patient peripheral blood samples obtained at diagnosis, as determined by droplet digital PCR (ddPCR). Phenotypically normal cell populations were isolated by fluorescence activated cell sorting according to the strategy depicted in Fig. 2a. Mutant allele frequency ~50% is consistent with a heterozygous cell population. Departures from 50% mutant allele frequency may be stochastic51, related to clonal heterogeneity, or due to the presence of copy number variations, for example loss of the wild type allele (loss of heterozygosity) or amplification of the mutant allele. NA, no cell population detected; HSC, haematopoietic stem cell; MPP, multipotent progenitor; CMP, common myeloid progenitor; MEP, megakaryocyte erythroid progenitor; MLP, multilymphoid progenitor; GMP, granulocyte monocyte progenitor; NK, natural killer cells. Blank boxes indicate no DNMT3A or NPM1 mutation detected. b, Representative plots showing ddPCR analysis of DNMT3Amut and NPM1c allele frequency in sorted cell populations from patient no. 11. The mutant allele frequency (%) is indicated on each plot.