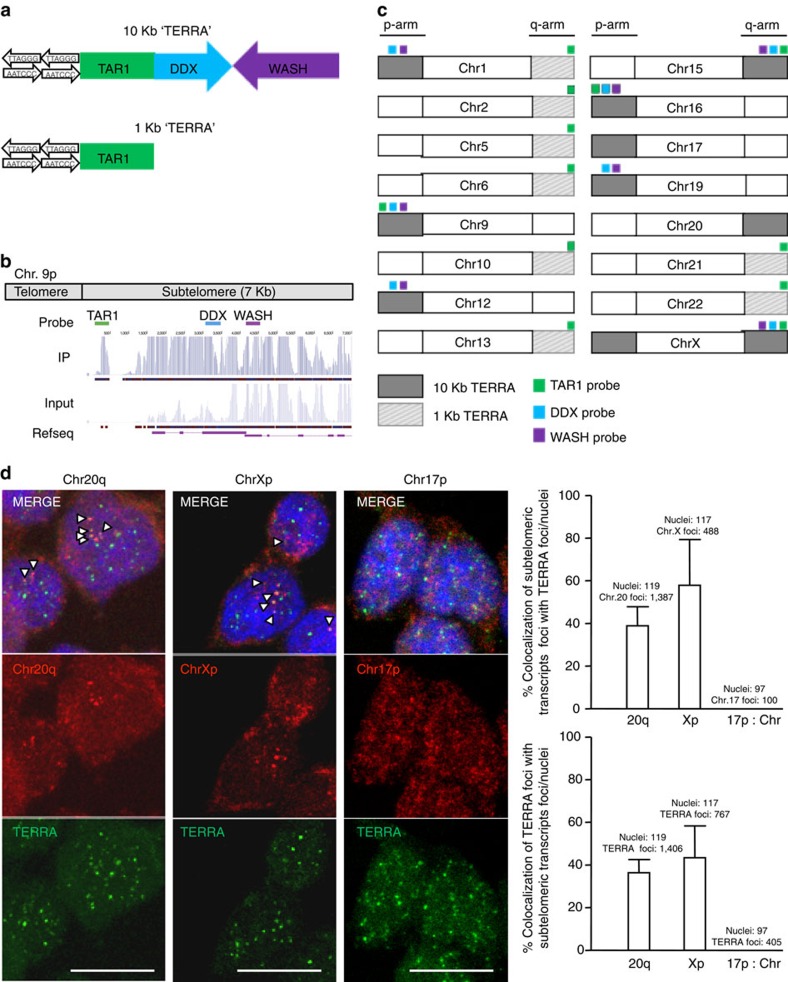
Figure 1. Transcripts arising from the subtelomere of chromosomes 20q and Xp co-localize with TERRA.
(a) Schematic of the three motifs in the structurally conserved TERRA loci proposed in Porro et al.,13, the repetitive region TAR1 and the two lncRNA families, DDX11L and WASH. (b) Example of the position of the probes against the structurally conserved motifs in the subtelomere of chromosome 9p. The alignment of the RNA-seq reads obtained in a TERRA-IP compared with the input13 and an annotated Ref seq in this region is also shown. (c) Schematic of the position of TERRA loci proposed in Porro et al.,13 within all chromosomes. The 10 kb TERRA is coloured in grey and the 1-kb TERRA with stripes. The position of the probes against TAR1, DDX11L and WASH is also indicated in different colours. (d) Confocal microscopy images of double RNA-FISH using probes targeting either subtelomere 20q, Xp or 17p transcripts (red) and TERRA's telomeric tract (green). The representative images are the merge of four individual confocal images. Co-localization events were only counted as positive when detected in the individual confocal images. Arrowheads indicate real co-localization events detected in the individual confocal images. (Top graph) The percentage of co-localization of either subtelomere 20q foci or subtelomere Xp foci with TERRA foci with respect the total number of 20q or Xp foci is represented (mean±s.d., n=number of nuclei). Total number of nuclei and foci are also indicated. (Bottom graph) The percentage of co-localization of TERRA foci with either subtelomere 20q foci or subtelomere Xp foci with respect the total number of TERRA foci is also represented (mean±s.d., n=number of nuclei). Total number of nuclei and foci are also indicated. Scale bar, 10 μm.