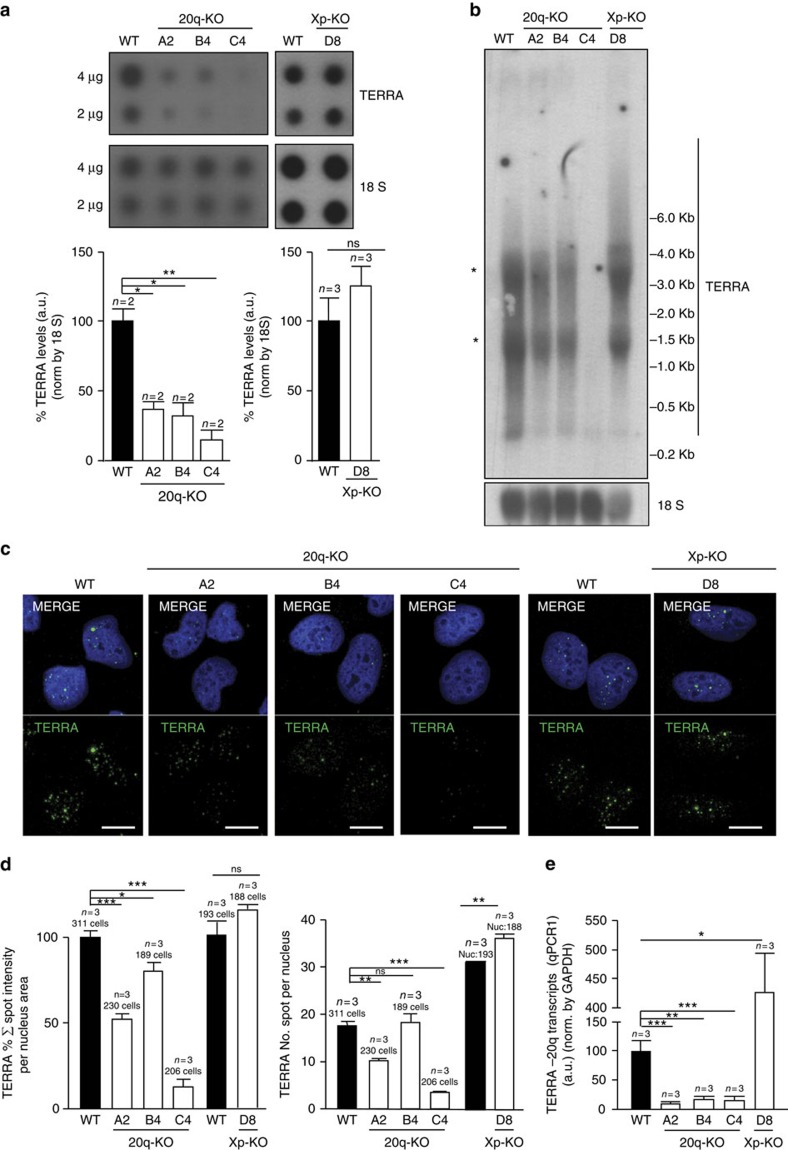
Figure 3. Deletion of the TERRA-20q locus dramatically affects TERRA expression.
(a) RNA from the U2OS cells WT or from the three 20q-KO clones (A2, B4 and C4) and the Xp-KO (D8) clone was isolated and used for TERRA detection by RNA dot-blot with a probe against the TERRA-UUAGGG-tract; 18S serves as loading control. (Graph) TERRA quantification normalized by 18S (mean values±s.e.m.). (b) Northern blotting using 32P-dCTP-labelled probe against TERRA-UUAGGG-tract in the U2OS cells WT or KO for the 20q or Xp loci. 18S was included as a loading control. *Unspecific band due to cross-hybridization with rRNA 18S and 28S. (c) Representative confocal microscopy images of RNA-FISH against TERRA-UUAGGG-tract (green) in the U2OS cells WT and KO for the 20q (clones A2, B4 C4) and the Xp (clone D8) loci. Scale bar, 10 μm. (d) The graphs show (left) the quantification of the total spot intensity per nucleus normalized by nucleus area, (right) the total number of spots per nucleus in the three 20q-KO clones and in the Xp-KO clone D8 (mean values±s.e.m., n=3 independent experiments). (e) Detection of the 20q-TERRA transcripts by qPCR (primers were designed in the subtelomeric region). The percentage of enrichment of the 20q-TERRA transcripts in WT and in the 20q-KO clones (clones A2, B4 and C4) and in the Xp-KO (clone D8) normalized by GAPDH is shown. One-way Anova with Dunnett's post test was used for the statistical analysis of the 20q clones and the Student's t-test for the Xp clone (*P<0.05, **P<0.01 and ***P<0.001).