Summary
Microbial biotransformations are major contributors to the arsenic biogeocycle. In parallel with transformations of inorganic arsenic, organoarsenicals pathways have recently been recognized as important components of global cycling of arsenic. The well-characterized pathway of resistance to arsenate is reduction coupled to arsenite efflux. Here, we describe a new pathway of arsenate resistance involving biosynthesis and extrusion of an unusual pentavalent organoarsenical. A number of arsenic resistance (ars) operons have two genes of unknown function that are linked in these operons. One, gapdh, encodes the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase. The other, arsJ, encodes a major facilitator superfamily (MFS) protein. The two genes were cloned from the chromosome of Pseudomonas aeruginosa. When expressed together, but not alone, in Escherichia coli, gapdh and arsJ specifically conferred resistance to arsenate and decreased accumulation of As(V). Everted membrane vesicles from cells expressing arsJ accumulated As(V) in the presence of purified GAPDH, D-glceraldehylde 3-phosphate (G3P) and NAD+. GAPDH forms the unstable organoarsenical 1-arseno-3-phosphoglycerate (1As3PGA). We propose that ArsJ is an efflux permease that extrudes 1As3PGA from cells, where it rapidly dissociates into As(V) and 3-phosphoglycerate (3PGA), creating a novel pathway of arsenate resistance.
Introduction
The toxic metalloid arsenic is ubiquitous in the environment, coming from both natural and anthropogenic sources (Hughes et al., 2011). Exposure to high levels of arsenic causes acute toxicity, but chronic lower level exposure is related to carcinogenesis and other health effects (Mandal and Suzuki, 2002). Environmental arsenic occurs mainly as inorganic species, and association with minerals or organic substances in soils limits its mobility (Akter et al., 2005). Inorganic trivalent As(III) is considerably more toxic than pentavalent As(V), in large part because the trivalent species reacts with small molecule thiols and protein sulfhydryl groups, thereby affecting their function (Hughes et al., 2011). The lower toxicity of arsenate results primarily from its chemical similarity to phosphate. It acts as a phosphate analog in phosphorylation reactions, but the products are less stable and dissociate rapidly (Long and Ray, 1973; Byers et al., 1979; Moore et al., 1983).
As a result of continual exposure to arsenic, most organisms, from bacteria to humans, have evolved arsenic detoxification pathways (Rosen, 2002). In bacteria arsenate is taken up by phosphate transport systems (Rosenberg et al., 1977), where it is reduced to arsenite by arsenate reductases (Mukhopadhyay and Rosen, 2002). Subsequent arsenic resistance mechanisms use arsenite as a substrate for efflux (Yang et al., 2012) or methylation (Ajees and Rosen, 2015). It appears counterintuitive to convert the less toxic pentavelent inorganic arsenic to the more toxic trivalent species for detoxification. We have postulated that the detoxification pathways evolved when the atmosphere was neutral, and most arsenic was present as reduced As(III) (Liu et al., 2013). Only an As(III) efflux system would be required for resistance, and so nearly every extant prokaryote has one of two unrelated As(III) efflux permeases, either ArsB or Acr3 (Yang et al., 2012). Once the atmosphere became oxidizing, arsenic would have become predominately pentavalent As(V). At least three families of arsenate reductase enzymes arose, apparently by convergent evolution, that would transform As(V) to As(III), the substrate of the efflux permeases (Mukhopadhyay and Rosen, 2002).
Yet an arsenate efflux system would have several advantages. First, it would not require cellular reductants or reductases. Second, it would not require biotransformation to more reactive and toxic trivalent species. However, to date, no As(V) efflux systems have been observed. Here, we report the existence of a two-gene system encoded by an ars operon in P. aeruginosa DK2 that creates, in effect, an arsenate efflux pathway. The first gene, gapdh, encodes the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase. The other gene, which we have named arsJ, encodes a putative MFS transporter. When gapdh or arsJ were co-expressed, cells were resistant to As(V) but not As(III) or methylarsenate (MAs(V)) or methylarsenite (MAs(III)). Those cells accumulated less As(V) than control cells without the two genes, suggesting either reduced uptake or increased efflux of an arsenate-containing compound. Everted membrane vesicles prepared from cells expressing arsJ accumulated As(V) but not As(III) only when purified rabbit GAPDH and its substrates were present. These results indicate that ArsJ transports an arsenate-containing product of the GAPDH reaction. GAPDH catalyzes formation of 1-arseno-3-phosphoglycerate (1As3PGA), a highly unstable organoarsenical with a half-life of less than 2.5 s (Byers et al., 1979). We predict that 1As3PGA is the true substrate of ArsJ and propose that in vivo GAPDH forms 1As3PGA, which is extruded from cells by ArsJ, conferring arsenate resistance. This novel coupling of a glycolytic enzyme and an organoarsenical efflux permease is the first identified transport pathway for arsenate resistance.
Results
Analysis of the ars operons of P. aeruginosa DK2
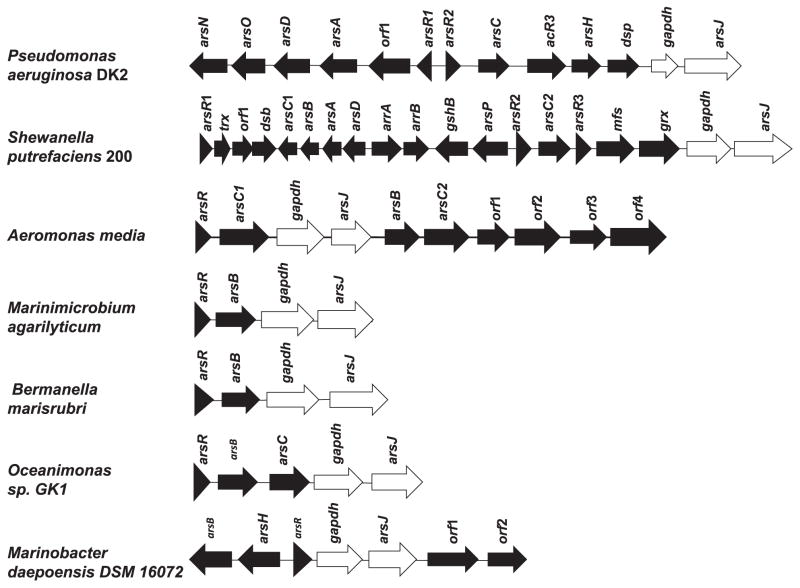
Arsenic is the most prevalent environmental toxin. As a result of its ubiquity, nearly every prokaryote and archaea has an ars operon. A number of ars operons include a pair of genes, gapdh and arsJ, with no known function in arsenic resistance. Representative examples are shown in Fig. 1. For example, in the chromosome of P. aeruginosa DK2 (accession number NC_018080.1) there are two ars operons from nucleotide 6068254 to nucleotide 6079515, a six-gene operon with arsR1-orf1-arsA-arsD-arsO-arsN and a divergently transcribed seven-gene ars operon with arsR2-arsC-acr3-arsH-dspgapdh-arsJ. Some of the gene products have known functions in arsenic resistance. The ArsRs are As(III)-responsive transcriptional repressors (Shi et al., 1994; Qin et al., 2007; Ordóñez et al., 2008). ArsD is a metallochaperone that transfers As(III) to the ArsA As(III) ATPase (Lin et al., 2006). Acr3 is an As(III) efflux permease that can interact with ArsA to form an ATP-driven efflux pump (Fu et al., 2009). ArsC is an arsenate reductase (Mukhopadhyay and Rosen, 2002). ArsH is a MAs(III) oxidase (Chen et al., 2015). The roles of Orf1, ArsO, ArsN and Dsp (dual specific protein phosphatase) in arsenic resistance are not known.
Fig. 1.
The gapdh and arsJ genes are linked in ars operons. Shown are representative ars operons containing gapdh and arsJ genes (white fill). Pseudomonas aeruginosa DK2 (accession number NC_018080), Shewanella putrefaciens 200 (accession number CP002457), Aeromonas media (accession number CP007567), Marinimicrobium agarilyticum (accession number AUHU00000000), Bermanella marisrubri (accession number AAQH01000000), Oceanimonas sp. GK1 (accession number CP003171), Marinobacter daepoensis (ATWI01000000).
In this study, we define the roles of the gapdh and arsJ gene products. The gapdh gene product (accession number WP_003109848.1) is a typical glyceraldehyde 3-phosphate dehydrogenase (GAPDH), the cytosolic NAD+-dependent enzyme (EC 1.2.1.12) found in all organisms so far studied (Fothergill-Gilmore and Michels, 1993). Representative examples are shown in Supporting Information Fig. 1S. The arsJ gene product (accession number WP_003109849.1) appears to be a novel 410-residue MFS membrane protein. The transmembrane topology of P. aeruginosa DK2 ArsJ was analyzed using the the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), which predicts 10 transmembrane spanning segments. In the NCBI database there are 2336 entries for ArsJ sequences, primarily in gammaproteobacteria. We analyzed representative examples (Supporting Information Fig. 2S), and each was in an ars operon adjacent to a gapdh gene (Fig. 1). There are no related sequences in archaea. A number of eukaryotic algae have related genes. In the alga Chlamydomonas reinhardtii, the gene for ArsJ (accession number XP_001702048.1) is also adjacent to genes for GAPDH (accession number XP_001702068.1) and phosphoglycerate kinase (accession number XP_001702049.1). There are also genes for related MFS proteins in the mycorrhizal fungus Rhizophagus irregularis DAOM 181602 (accession number ESA15517.1) and in the acorn worm Saccoglossus kowalevskii (accession number NP_001171775.1), but neither appears to be adjacent to a gapdh gene, and functional relatedness of the gene products to ArsJ is uncertain.
GAPDH and ArsJ synergistically confer As(V) resistance
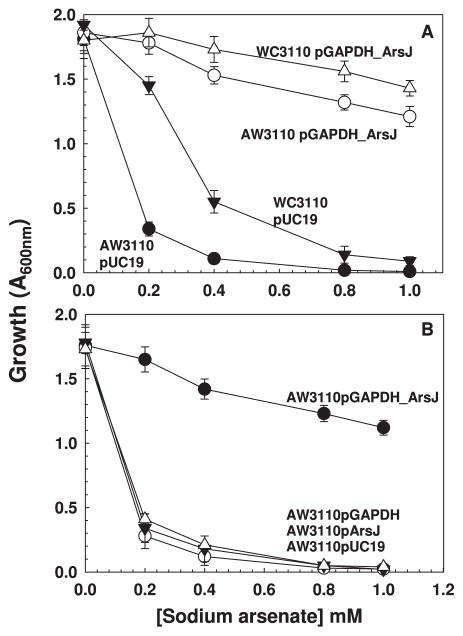
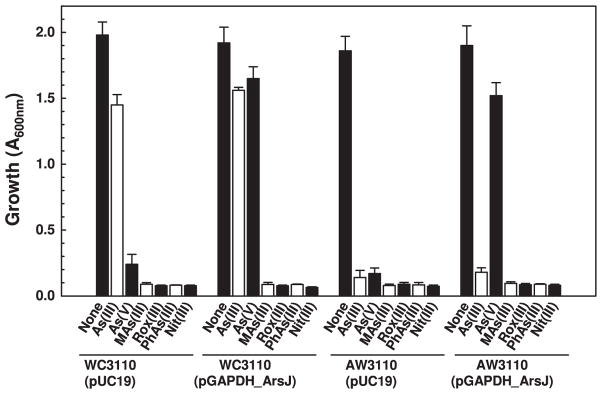
The physical association of the gapdh and arsJ genes in ars operon suggests a common function. For example, the arsD and arsA genes are nearly always adjacent to each other in ars operons, and ArsD serves as an As(III) chaperone for the ArsA As(III)-translocating ATPase (Lin et al., 2006). The gapdh and arsJ genes were cloned individually and together into pUC19 under control of the lac promoter, creating plasmids pGAPDH, pArsJ and pGAPDH_ArsJ, which were expressed in either in the arsenate sensitive E. coli strain WC3110 (ΔarsC) (Mukhopadhyay et al., 2000) or the arsenic-hypersensitive strain AW3110 (ΔarsRBC) (Carlin et al., 1995). Resistance to inorganic and organic arsenicals was assayed in these strains. Cells of either strain AW3110 or WC3110 with plasmid pGAPDH_ArsJ were resistant to As(V) compared with the same strain with vector only (Fig. 2A). Cells of strain AW3110 with either pGAPDH or pArsJ remained As(V) sensitive (Fig. 2B), as were cells of WC3110 with the individual genes (data not shown). Cells of AW3110 with both genes was also sensitive to As(III), but WC3110 with plasmid pGAPDH_ArsJ is resistant because it still has an arsB gene. Cells of either strain expressing both genes remained sensitive to the trivalent organoarsenicals MAs(III), PhAs(III), Rox(III) and Nit(III) compared with the same strain with vector only (Fig. 3). These results clearly demonstrate that both the gapdh and arsJ genes synergistically and specifically detoxify inorganic As(V).
Fig. 2. GAPDH and PaArsJ confer synergistic resistance to arsenate.
A. Arsenate resistance in E. coli strain AW3110 (ΔarsRBC) (▼,Δ) or WC3110 (ΔarsC) (●,○) bearing the indicated plasmids was assayed as described in Experimental Procedures. (▼,●), vector plasmid pUC19; (Δ,○), pGAPDH_ArsJ.
B. Arsenate resistance in E. coli strain AW3110 with (●) pGAPDH_ArsJ, (Δ), pGAPDH, (▼) pArsJ, (○), pUC19. Growth was estimated from the absorption at 600 nm after 24 h at 37°C. Data are the mean±SE (n=3).
Fig. 3.
Co-expression of gapdh and arsJ confer resistance to arsenate. Overnight cultures of E. coli strain WC3110 or AW3110 bearing either pGAPDH_ArsJ or vector plasmid pUC19 were diluted 100-fold into fresh low phosphate medium containing 0.1 mM As(III), 0.8 mM As(V), 10 μM MAs(III), 0.5 μM PhAs(III), 2 μM Rox(III) or 2 μM Nit(III), as indicated. Expression of the gapdh and arsJ genes was induced with 0.3 mM IPTG. Growth was estimated from the absorption at 600 nm after 24 h at 37°C. Data are the mean±SE (n=3).
GAPDH and ArsJ synergistically reduce cellular As(V) accumulation
ArsJ is predicted to be an MFS membrane protein, suggesting that it could be a permease. The effect of expression of gapdh and arsJ on accumulation of As(V), As(III) and MAs(III) was examined (Fig. 4). Cells expressing both gapdh and arsJ accumulated considerably less As(V) compared with cells lacking the two genes. However, there was little difference in uptake of either As(III) or MAs(III) between cells expressing the two genes or vector only. Cells with vector only did not accumulate MAs(V) (data not shown), so it was not possible to determine whether cells of E. coli AW3110 expressing the two genes can extrude the pentavalent species. These results indicate that the combination of GAPDH and ArsJ reduce accumulation of As(V). This could be the result of decreased uptake or increased efflux of inorganic As(V) or a compound containing As(V).
Fig. 4.
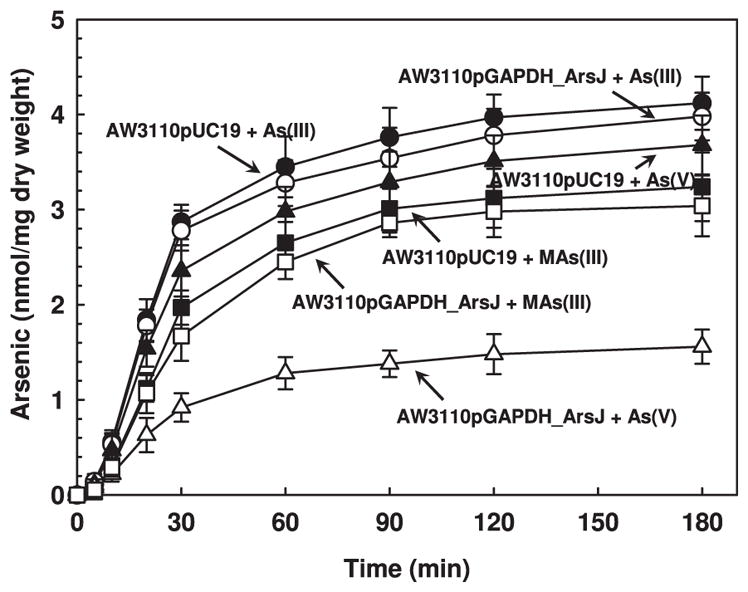
GAPDH and ArsJ reduce cellular As(V) accumulation. Uptake of arsenicals by E. coli AW3110 carrying either vector plasmid pUC19 (filled symbols) or pGAPDH_ArsJ (open symbols) was assayed with 20 μM, final concentration, of the indicated arsenicals. (Δ,▲) As(V), (○,●), As(III), (□,■) MAs(III), was performed as described in Experimental procedures. Data are the mean±SE (n=3).
ArsJ transports 1-arseno-3-phosphoglycerate
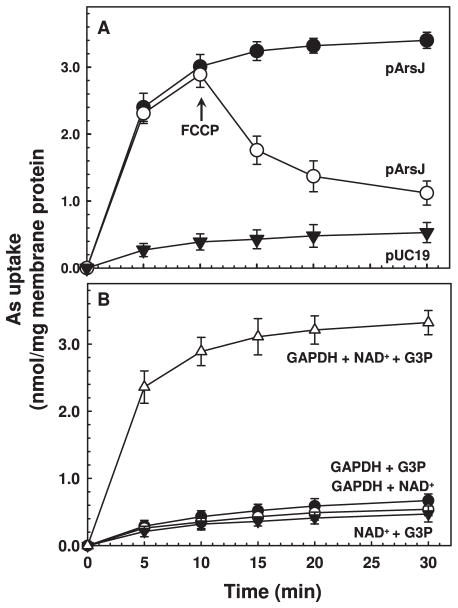
To distinguish between reduced uptake or increased efflux, transport into everted membrane vesicles prepared from cells of E. coli AW3110pArsJ was measured. Uptake into everted membrane vesicles is the equivalent of efflux from cells (Rosen and Tsuchiya, 1979). Vesicle transport was assayed in the presence of purified rabbit GAPDH, G3P and NAD+. Cells expressing arsJ accumulated As(V), while cells with vector only had considerably less uptake (Fig. 5A). Accumulated As(V) was released by addition of the uncoupler carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), indicating that uptake is energy dependent. The usual energy source in everted vesicle transport is NADH, which generates a protonmotive force through respiration. We assume that GAPDH generates sufficient NADH to drive the transport reaction.
Fig. 5. Uptake of 1-arseno-3-phosphoglycerate in everted membrane vesicles.
A. Accumulation of 1As3PGA in everted membrane vesicles prepared from E. coli strain AW3110 harboring either plasmid pArsJ (●,○) or vector plasmid pUC19 (▼) was assayed as described in Experimental procedures using 1 mg/ml of membrane protein. To produce 1AS3PGA in situ, the reaction mixture contained, 5 units of commercial rabbit GAPDH, 10 mM DTT, 5 mM NAD+ and 5 mM As(V). The reaction was initiated by addition of 5 mM G3P, final concentration. 10 μM FCCP, final concentration, was added at the indicated time (arrow).
B. The requirements for in situ generation of 1As3PGA were determined by addition of the components of the GAPDH reaction. (Δ), GAPDH, NAD+ and G3P; (●), GAPDH and G3P; (○), GAPDH and NAD+; (▼), NAD+ and G3P. Vesicular arsenic was determined by ICP-MS. Data are the mean±SE (n=3).
What is the chemical nature of the substrate for ArsJ? As(V) uptake was observed only when purified GAPDH, G3P and NAD+ were present (Fig. 5B). Little uptake occurred in the absence of GAPDH, G3P or NAD+. Thus the GAPDH reaction is required for As(V) accumulation in vesicles. In the presence of As(V), the product of the reaction is 1As3PGA (Byers et al., 1979), and it is reasonable to assume that 1As3PGA is the true substrate of ArsJ. 1As3PGA has a half-life of less than 2.5 s, so it must be generated in situ. In addition, ArsJ is specific; there was little uptake when As(V) was substituted with As(III), MAs(III), MAs(V) or DMAs(V), which are not substrates of GAPDH (Fig. 6).
Fig. 6. ArsJ substrate specificity. Transport assays in everted membrane vesicles prepared from E. coli strain AW3110pArsJ were performed as described in Experimental procedures.
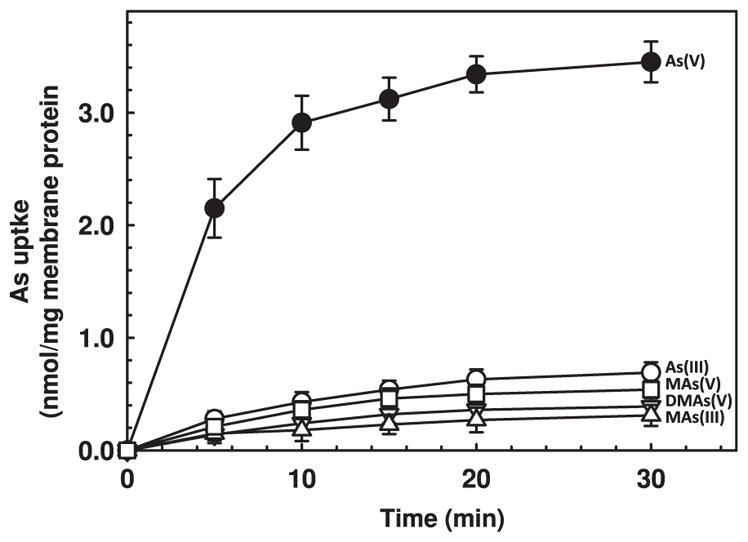
Arsenicals were added at 5 mM, final concentration. (●), As(V); (○), As(III); (□), MAs(V); (▽), DMAs(V); (Δ), MAs(III). Vesicular arsenic was determined by ICP-MS. Data are the mean±SE (n=3).
GAPDH activity with arsenate
GAPDH activity was assayed by NADH formation (Supporting Information Fig. 3S). No activity was observed in the absence of inorganic arsenate (Supporting Information Fig. 3SA) or in the presence of As(III), MAs(III), MAs(V) or DMAs(V) (Supporting Information Fig. 4SA). GAPDH activity required G3P, NAD+ and As(V) (Supporting Information Fig. 4SB) and was dependent on the arsenate concentration, with an apparent Km of 0.46 mM (Supporting Information Fig. 5S). GAPDH has been reported to reduce As(V) to As(III) (Gregus and Nemeti, 2005; Nemeti et al., 2006). However, in our hands, no As(V) reduction was observed either in vivo in cells of E. coli expressing gapdh (Supporting Information Fig. 6SA) or in vitro with purified rabbit GAPDH (Supporting Information Fig. 6SB). Cells of neither E. coli AW3110 nor WC3110, both of which have the arsC gene deleted, reduced significant amounts of As(V). Under the same conditions, the parental strain W3110, which has a chromosomal arsC gene, reduced more than half of the As(V) to As(III). Similarly, there was nearly no reduction of arsenate by purified GAPDH even after 40 min. Thus, our results cannot be explained by any adventitious arsenate reductase activity of GAPDH.
When both arsenate and phosphate were assayed at 50 μM, final concentration, the steady state rate of GAPDH activity with phosphate was very low compared with the same concentration of arsenate (Supporting Information Fig. 3SB). Activity was very rapid at phosphate concentrations in the range of 3–25 mM, too fast to acquire initial rates with our plate reader (Supporting Information Fig. 3SC), but the reaction reached a steady state within the first few seconds. The difference in GADPH reactivity with arsenate or phosphate may relate to the fact that the two products have three orders of magnitude difference in stability (t1/2 of <2.5 s for 1As3PGA versus 48 min for 1,3-PGA) (Byers et al., 1979). 1,3-PGA builds up and becomes inhibitory, while 1As3PGA never accumulates, which is the basis of the uncoupling effect of arsenate. Thus GAPDH activity correlates with ArsJ activity. These results are consistent with generation of 1As3PGA by GAPDH coupled with transport by ArsJ.
Discussion
Arsenite is more toxic than arsenate. Arsenite inhibits many cysteine-containing enzymes and bind to thiol-containing metabolites such as reduced glutathione and lipoic acid. Arsenate exerts its toxicity as a phosphate analog, where it substitutes for Pi in phosphorylation reactions. Since the arsenylated products are unstable, As(V) uncouples substrate-level phosphorylation reactions in glycolysis (Pillai, 1938) and ATP production in oxidative phosphorylation (Azzone and Ernster, 1961). Yet nearly every microbial protein involved in resistance to arsenicals uses the trivalent species as a ligand or substrate, including the As(III)-responsive transcriptional repressor ArsR (Shi et al., 1994), the As(III) metallochaperone ArsD (Lin et al., 2006), the ArsA As(III) ATPase (Rosen et al., 1988), the ArsB and Acr3 As(III) efflux permeases (Meng et al., 2004; Villadangos et al., 2012), the ArsM As(III) S-adenosylmethionine methyltransferase (Qin et al., 2006). The most obvious exceptions are the arsenate reductases, which reduce As(V) to As(III) in order to become the substrates of the resistance mechanisms (Mukhopadhyay and Rosen, 2002). The ArrAB respiratory arsenate reductase also transforms the pentavelent species, but for energy production and not resistance (Saltikov and Newman, 2003).
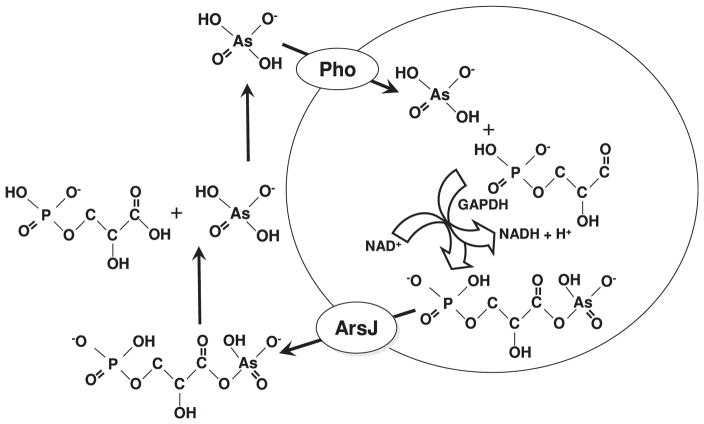
Yet no arsenate efflux carriers or pumps have been identified. In this study, we identify a two-step reaction that creates a pathway for arsenate efflux. There are a number of genes with unknown function in a wide variety of ars operons. We began with the assumption that all genes in ars operons encode an arsenic-related function. We noted that various ars operons have two genes, gapdh, which encodes a typical glyceraldhyde-3-phosphate dehydrogenase or GAPDH enzyme, and a second encoding an MFS membrane protein that we have named arsJ. GAPDH is an NAD+-dependent enzyme present in nearly every organism. This cytosolic enzyme plays a major role in glycolysis and gluconeogenesis. The substrates for the glycolytic direction is inorganic phosphate, D-glyceraldehyde-3-phosphate and NAD+ and the product is 1,3-bisphosphoglycerate. As G3P is a metabolite in glycolysis, it is always present during glucose metabolism. Arsenate can substitute for phosphate, with production of the product 1As3PGA, which is unstable and immediately hydrolyzes to inorganic arsenate and 3PGA, creating a futile cycle that uncouples glycolysis (Byers et al., 1979). ArsJ is an MFS membrane protein with a likely transport function. Since the genes for ArsJ and GAPDH are linked, it seemed reasonable to consider that the ArsJ could be an efflux system for 1As3PGA (Fig. 7). Whether 1,3-PGA is also a substrate was not tested, but it seems unlikely that the function of a gene in an ars operon would be to deplete cells of phosphate. Even if it is specific for arsenate, it still creates a potential futile cycle of arsenate uptake and efflux, but the data in Fig. 4 suggests that it functions more like a bilge pump. With a half life of <2.5 s for 1As3PGA, the organoarsenical is sufficiently stable to be transported by ArsJ before it hydrolyzes. Formation of 1As3PGA with intracellular As(V) by GAPDH coupled with extrusion by ArsJ and hydrolysis to extracellular As(V) creates the equivalent of an As(V) efflux system. Although 1As3PGA is not available to add as a substrate, it is generated in situ by adding purified GAPDH and the reaction components to an in vitro vesicle transport assay. There does not appear to be anything special about the sequence of the ars GAPDH that differentiates it from glycolytic GAPDH enzymes, so why does not gapA gene product of E. coli AW3110 or WC3110 produce sufficient 1As3PGA to confer arsenate resistance or efflux? It is likely the result of high levels of gapdh transcription from the strong ars promoter under natural conditions or from the lac promoter in the recombinant plasmid used in this study. The gapA gene of E. coli is under control of multiple promoters but is most highly expressed at the beginning of exponential phase or during gluconeogenesis (Charpentier and Branlant, 1994). Under the conditions of this study the ars gapdh gene is probably much more highly expressed than the chromosomal gapA gene. In summary, the ars operon genes gapdh and arsJ encode a new and novel pentavalent organoarsenical resistance, the only identified pathway for efflux of arsenate.
Fig. 7.
Model of synergy between GAPDH and ArsJ in arsenate transport and detoxification. Arsenate is taken up by phosphate transporters. GAPDH catalyzes arsenylation of G3P to form 1As3PGA, which is the substrate of the ArsJ efflux permease. Extracellular 1As3PGA is unstable and spontaneously hydrolyzes into As(V) and 3PGA.
Experimental procedures
Strains, plasmids, medium and reagents
Escherichia coli Stellar™ (Clontech Laboratories, Mountain View, CA; F−, endA1, supE44, thi-1, recA1, relA1, gyrA96 phoA, Φ80d lacZΔ M15, Δ(lacZYA-argF)U169, Δ(mrrhsdRMS-mcrBC), ΔmcrA, λ−) was used for plasmid DNA construction and replication. E. coli AW3110(DE3) (ΔarsRBC::cam F–IN(rrn-rrnE) (Carlin et al., 1995), which is hypersensitive to As(III), and WC3110(DE3) (ΔarsC) (Sundaram et al., 2008) were used for complementation studies. For most experiments, cultures of E. coli bearing the indicated plasmids were grown aerobically in Luria–Bertani (LB) medium (Sambrook et al., 1989) or low phosphate medium (Oden et al., 1994) at 30 or 37°C supplemented with 125 μg/ml ampicillin or 34 μg/ml chloramphenicol, as indicated. Bacterial growth was monitored by measuring the absorbance at 600 nm (A600). All reagents were obtained from commercial sources. Rabbit GAPDH (CAS: 9001-50-7) and G3P (CAS 591-57-1) were obtained from Sigma-Aldrich (St Louis, MO). Roxarsone (CAS 121-19-7) was obtained from ThermoFisher Acros Organics Division (Waltham, MA). Monomethylarsonic acid (MAs(V)) was obtained from Chem Service (West Chester, PA, USA). MAs(III), Rox(III), Nit(III) were prepared by reduction of the pentavalent forms (Reay and Asher, 1977).
Plasmid construction
For expression of GAPDH (accession number WP_003109848) and ArsJ (accession number WP_003109849) in E. coli, the gapdh and arsJ genes were cloned individually and together from P. aeruginosa DK2 genomic DNA (a gift from Søren Molin, Technical University of Denmark) into plasmid pUC19 under control of the lac promoter, creating plasmids pGAPDH, pArsJ and pGAPDH_ArsJ. The forward and reverse primers for cloning the gapdh and arsJ genes together were: 5′-CCCAAGCTTATGGCCATCAAAGTAGGCATC-3′ (HindIII site underlined) and 5′-ACGCGTCGACTCAGTTCGGGGTGGTCACCAC-3′ (SalI site underlined), respectively. The PCR fragment was gel purified and digested using the underlined restriction enzymes, and ligated into vector plasmid pUC19, which was also digested with HindIII and SalI, generating plasmid pGAPDH_ArsJ. To clone the gapdh gene alone, the forward and reverse primers were 5′-CCCAAGCTTATGGCCATCAAAGTAGGCATC-3′ (HindIII site underlined) and 5′-ACGCGTCGACTCAGCCAGCCAGGCCGACCAG-3′ (SalI site underlined), respectively. To clone the arsJ gene alone, the forward and reverse primers were 5′-CCCAAGCTTATGAAGGCGCTATCGTCGCTCT-3′ (HindIII site underlined) and 5′-ACGCGTCGACTCAGTTCGGGGTGGTCACCAC-3′ (SalI site underlined), respectively.
Comparison of GAPDH and ArsJ sequences
Multiple alignment of GAPDH (Supporting Information Fig. 1S) and ArsJ (Supporting Information Fig. 2S) sequences was calculated with CLUSTAL W (Thompson et al., 1994).
Metalloid resistance assays
For metalloid resistance assays in liquid medium, competent cells of either AW3110 or WC3110, as indicated, were transformed with the indicated plasmids. Cells were grown overnight with shaking at 37°C in LB medium with 125 μg ml−1 ampicillin. Overnight cultures were diluted 100-fold in low phosphate medium containing various concentrations of either trivalent or pentavalent arsenicals plus 0.3 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and incubated at 37°C with shaking for another 24 h. Growth was estimated from the absorbance at 600 nm.
Metalloid transport assays in whole cells
For in vivo uptake assays, E. coli cells were grown in low phosphate medium at 37°C to a cell density (A600nm) of 2. The cells were harvested and suspended in 1/5th volume of a buffer solution consisting of 75 mM HEPES-KOH, pH 7.5, 0.15 M KCl and 1 mM MgSO4. To initiate the transport reaction, arsenicals were added at a final concentration of 20 μM to 1 ml of cell suspension. Portions (0.1 ml) were withdrawn at the indicated times, filtered through nitrocellulose filters (0.2 μm pore diameter; EMD Millipore, Billerica, MA) and washed twice at room temperature with 5 ml of the same buffer. The filters were digested with 0.3 ml of concentrated HNO3 (68–70%) overnight at room temperature. The dissolved filters were incubated for 10 min at 70°C, allowed to cool to room temperature and diluted with HPLC-grade water (Sigma-Aldrich, St. Louis) to produce a final HNO3 concentration of 2%. Arsenic was quantified by inductively coupled mass spectroscopy (ICP-MS). Standard solutions were made in the range of 0.5–50 ppb in 2% nitric acid using an arsenic standard (Ultra Scientific, N. Kingstown, RI).
Assay of GAPDH activity
GAPDH activity was assayed as described (Heinz and Freimuller, 1982). The reaction was initiated by addition of 0.6 mM G3P, final concentration, to a reaction mixture containing 25 mM Bis-Tris propane (pH 7.0), 0.6 mM NAD+, 10 mM DTT, 1 unit GAPDH and the indicated concentrations of inorganic phosphate or arsenate at 25°C. The absorbance at 340 nm was measured with a Synergy H4 Hybrid MultiMode microplate reader.
Metalloid uptake assays in everted membrane vesicles
Everted membrane vesicles and transport assays were performed as described (Villadangos et al., 2012). Transport assays were performed in a buffer consisting of 75 mM HEPES-KOH, pH 7.5, 0.15 M K2SO4, 1 mM MgSO4 and 0.25 M sucrose. The reaction mixture contained 1 mg ml−1 membrane proteins, 5 units of GAPDH, 10 mM DTT, 5 mM NAD+ and the indicated arsenicals at 5 mM, final concentration, in a final volume of 0.6 ml of the same buffer. The reaction was initiated by addition of 5 mM G3P. The uncoupler carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) was added as indicated. Portions (0.1 ml) were withdrawn at the indicated times, filtered through 0.2 μm pore size nitrocellulose filters and washed twice with 5 ml of the same buffer. The arsenic content was determined by ICP-MS, as described above.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R37 GM55425. LDG was supported on USDA NIFA Hispanic Serving Institutions Higher Education Grant 2008-38422-19209. We thank Professor Søren Molin for the gift of P. aeruginosa DK2 genomic DNA and Hiranmoy Bhattacharjee for advice.
Footnotes
Author Contributions: JC, MY and BPR made major contributions to the acquisition, analysis and interpretation of data and wrote the manuscript. LDG contributed to acquisition of the data.
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Ajees AA, Rosen BP. As(III) S-adenosylmethionine methyltransferases and other arsenic binding proteins. Geomicrobiol J. 2015;32:570–576. doi: 10.1080/01490451.2014.908983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akter KF, Owens G, Davey DE, Naidu R. Arsenic speciation and toxicity in biological systems. Rev Environ Contam Toxicol. 2005;184:97–149. doi: 10.1007/0-387-27565-7_3. [DOI] [PubMed] [Google Scholar]
- Azzone GF, Ernster L. Compartmentation of mitochondrial phosphorylations as disclosed by studies with arsenate. J Biol Chem. 1961;236:1510–1517. [PubMed] [Google Scholar]
- Byers LD, She HS, Alayoff A. Interaction of phosphate analogues with glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 1979;18:2471–2480. doi: 10.1021/bi00579a006. [DOI] [PubMed] [Google Scholar]
- Carlin A, Shi W, Dey S, Rosen BP. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol. 1995;177:981–986. doi: 10.1128/jb.177.4.981-986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier B, Branlant C. The Escherichia coli gapA gene is transcribed by the vegetative RNA polymerase holoenzyme E sigma 70 and by the heat shock RNA polymerase E sigma 32. J Bacteriol. 1994;176:830–839. doi: 10.1128/jb.176.3.830-839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bhattacharjee H, Rosen BP. ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol Microbiol. 2015;96:1042–1052. doi: 10.1111/mmi.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fothergill-Gilmore LA, Michels PA. Evolution of glycolysis. Prog Biophys Mol Biol. 1993;59:105–235. doi: 10.1016/0079-6107(93)90001-z. [DOI] [PubMed] [Google Scholar]
- Fu HL, Meng Y, Ordonez E, Villadangos AF, Bhattacharjee H, Gill JA, et al. Properties of arsenite efflux permeases (Acr3) from Alkaliphilus metalli-redigens and Corynebacterium glutamicum. J Biol Chem. 2009;284:19887–19895. doi: 10.1074/jbc.M109.011882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregus Z, Nemeti B. The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase works as an arsenate reductase in human red blood cells and rat liver cytosol. Toxicol Sci. 2005;85:859–869. doi: 10.1093/toxsci/kfi158. [DOI] [PubMed] [Google Scholar]
- Heinz F, Freimuller B. Glyceraldehyde-3-phosphate dehydrogenase from human tissues. Methods Enzymol. 1982;89(Pt D):301–305. doi: 10.1016/s0076-6879(82)89054-x. [DOI] [PubMed] [Google Scholar]
- Hughes MF, Beck BD, Chen Y, Lewis AS, Thomas DJ. Arsenic exposure and toxicology: a historical perspective. Toxicol Sci. 2011;123:305–332. doi: 10.1093/toxsci/kfr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YF, Walmsley AR, Rosen BP. An arsenic metallochaperone for an arsenic detoxification pump. Proc Natl Acad Sci USA. 2006;103:15617–15622. doi: 10.1073/pnas.0603974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Rensing C, Rosen BP. Resistance pathways for metalloids and toxic metals. In: Culotta V, Scott RA, editors. Metals in Cells. Hoboken, NJ: Wiley and Sons, Inc; 2013. pp. 429–442. [Google Scholar]
- Long JW, Ray WJ., Jr Kinetics and thermodynamics of the formation of glucose arsenate. Reaction of glucose arsenate with phosphoglucomutase. Biochemistry. 1973;12:3932–3937. doi: 10.1021/bi00744a023. [DOI] [PubMed] [Google Scholar]
- Mandal BK, Suzuki KT. Arsenic round the world: a review. Talanta. 2002;58:201–235. [PubMed] [Google Scholar]
- Meng YL, Liu Z, Rosen BP. As(III) and Sb(III) uptake by GlpF and efflux by ArsB in Escherichia coli. J Biol Chem. 2004;279:18334–18341. doi: 10.1074/jbc.M400037200. [DOI] [PubMed] [Google Scholar]
- Moore SA, Moennich DM, Gresser MJ. Synthesis and hydrolysis of ADP-arsenate by beef heart submitochondrial particles. J Biol Chem. 1983;258:6266–6271. [PubMed] [Google Scholar]
- Mukhopadhyay R, Rosen BP. Arsenate reductases in prokaryotes and eukaryotes. Environ Health Perspect. 2002;110(Suppl 5):745–748. doi: 10.1289/ehp.02110s5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Shi J, Rosen BP. Purification and characterization of Acr2p, the Saccharomyces cerevisiae arsenate reductase. J Biol Chem. 2000;275:21149–21157. doi: 10.1074/jbc.M910401199. [DOI] [PubMed] [Google Scholar]
- Nemeti B, Csanaky I, Gregus Z. Effect of an inactivator of glyceraldehyde-3-phosphate dehydrogenase, a fortuitous arsenate reductase, on disposition of arsenate in rats. Toxicol Sci. 2006;90:49–60. doi: 10.1093/toxsci/kfj058. [DOI] [PubMed] [Google Scholar]
- Oden KL, Gladysheva TB, Rosen BP. Arsenate reduction mediated by the plasmid-encoded ArsC protein is coupled to glutathione. Mol Microbiol. 1994;12:301–306. doi: 10.1111/j.1365-2958.1994.tb01018.x. [DOI] [PubMed] [Google Scholar]
- Ordóñez E, Thiyagarajan S, Cook JD, Stemmler TL, Gil JA, Mateos LM, Rosen BP. Evolution of metal(loid) binding sites in transcriptional regulators. J Biol Chem. 2008;283:25706–25714. doi: 10.1074/jbc.M803209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RK. Action of arsenate in glycolysis. Biochem J. 1938;32:1961–1973. doi: 10.1042/bj0321961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci USA. 2006;103:2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Fu HL, Ye J, Bencze KZ, Stemmler TL, Rawlings DE, Rosen BP. Convergent evolution of a new arsenic binding site in the ArsR/SmtB family of metalloregulators. J Biol Chem. 2007;282:34346–34355. doi: 10.1074/jbc.M706565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reay PF, Asher CJ. Preparation and purification of 74As-labeled arsenate and arsenite for use in biological experiments. Anal Biochem. 1977;78:557–560. doi: 10.1016/0003-2697(77)90117-8. [DOI] [PubMed] [Google Scholar]
- Rosen BP. Biochemistry of arsenic detoxification. FEBS Lett. 2002;529:86–92. doi: 10.1016/s0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- Rosen BP, Tsuchiya T. Preparation of everted membrane vesicles from Escherichia coli for the measurement of calcium transport. Methods Enzymol. 1979;56:233–241. doi: 10.1016/0076-6879(79)56026-1. [DOI] [PubMed] [Google Scholar]
- Rosen BP, Weigel U, Karkaria C, Gangola P. Molecular characterization of an anion pump. The arsA gene product is an arsenite(antimonate)-stimulated ATPase. J Biol Chem. 1988;263:3067–3070. [PubMed] [Google Scholar]
- Rosenberg H, Gerdes RG, Chegwidden K. Two systems for the uptake of phosphate in Escherichia coli. J Bacteriol. 1977;131:505–511. doi: 10.1128/jb.131.2.505-511.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltikov CW, Newman DK. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci USA. 2003;100:10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, a Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Shi W, Wu J, Rosen BP. Identification of a putative metal binding site in a new family of metalloregulatory proteins. J Biol Chem. 1994;269:19826–19829. [PubMed] [Google Scholar]
- Sundaram S, Rathinasabapathi B, Ma LQ, Rosen BP. An arsenate-activated glutaredoxin from the arsenic hyperaccumulator fern Pteris vittata L. regulates intracellular arsenite. J Biol Chem. 2008;283:6095–6101. doi: 10.1074/jbc.M704149200. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadangos AF, Fu HL, Gil JA, Messens J, Rosen BP, Mateos LM. Efflux permease CgAcr3-1 of Corynebacterium glutamicum is an arsenite-specific antiporter. J Biol Chem. 2012;287:723–735. doi: 10.1074/jbc.M111.263335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HC, Fu HL, Lin YF, Rosen BP. Pathways of arsenic uptake and efflux. Curr Top Membr. 2012;69:325–358. doi: 10.1016/B978-0-12-394390-3.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.