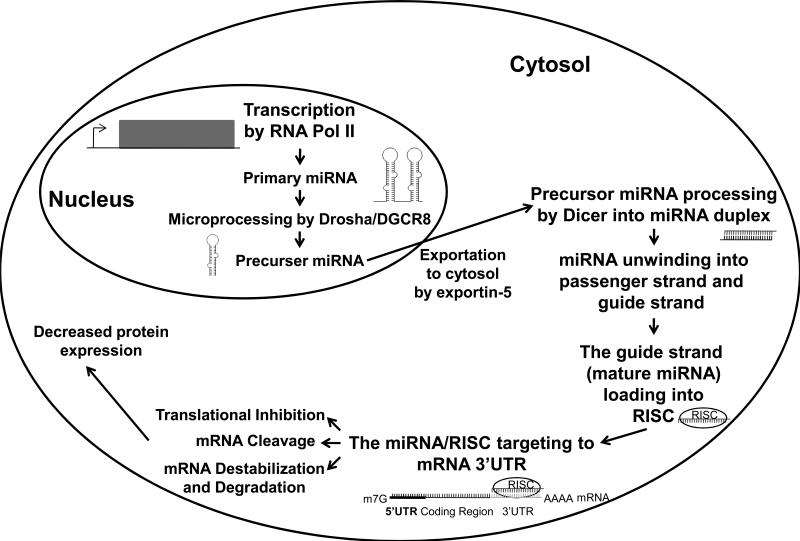
Figure 1. A brief schematic description of miRNA biogenesis and the mechanism of miRNA down-regulating protein-coding gene expression.
MiRNAs are transcribed in the nucleus by RNA Polymerase II (RNA Pol II) into primary-miRNA (pri-miRNA). The pri-miRNA is polyadenylated and capped, and then processed by Drosha and its binding partner DiGeorge syndrome critical region 8 (DGCR8) to generate the precursor-miRNA (pre-miRNA). This premiRNA is then exported out of the nucleus by exportin-5. In the cytosol, the pre-miRNA is processed via Dicer into a miRNA duplex. The miRNA duplex is then unwound into its guide strand (mature miRNA) and passenger strand (star strand). While the passenger strand is usually degraded, the mature miRNA then associates with the Argonaute protein and is subsequently loaded into the RNA-induced silencing complex (RISC). The mature miRNA/RISC complex goes to find its target mRNA based upon the miRNA seed sequence. The interaction of miRNA/RISC complex with the target mRNA can cause mRNA destabilization and degradation, cleavage, or inhibition of translation of the mRNA.