Abstract
Background
Patients with atopic dermatitis (AD) are prone to skin infections, with microbes like Staphylococcus aureus suspected of contributing to pathogenesis. Bleach baths might improve AD by reducing skin microbial burden.
Objective
To characterize the microbiota of lesional and nonlesional skin in young children with AD and controls and compare changes after treatment with a topical corticosteroid (TCS) alone or TCS + dilute bleach baths (BB).
Methods
In a randomized, placebo-controlled, single-blinded clinical trial in 21 children with AD and 14 healthy children, lesional and nonlesional AD skin was examined at baseline and after 4-week treatment with TCS alone or TCS plus BB. Microbial DNA was extracted for qPCR of predominant genera and 16S rRNA sequencing.
Results
At baseline, densities of total bacteria and Staphylococcus, including S. aureus, were significantly higher at the worst AD lesional site than nonlesional (p=0.001) or control (p<0.001) skin; bacterial communities on lesional and nonlesional AD skin significantly differed from each other (p=0.04) and from control (p<0.001). After TCS + BB or TCS alone, bacterial compositions on lesional skin normalized (p<0.0001), resembling nonlesional skin, with microbial diversity restored to control skin levels.
Limitations
The 4-week time period and/or the twice-weekly baths may not have been sufficient for additional impact on the cutaneous microbiome. More detailed sequencing may allow better characterization of the distinguishing taxa with BB-treatment.
Conclusions
Treatment with a topical corticosteroid cream suffices to normalize the cutaneous microbiota on lesional AD; after treatment, bacterial communities on lesional skin resemble nonlesional skin but remain distinct from control.
INTRODUCTION
Atopic dermatitis (AD) is a common, chronic inflammatory skin condition characterized by pruritic eczematous lesions in specific distribution patterns in infants and children.1 Morbidity in patients with AD is often due to cutaneous infections with particular bacteria, fungi, and viruses. Staphylococcus aureus is highly prevalent on AD skin, and culture-based studies have shown direct correlation between AD clinical severity and S. aureus density.2 Whether S. aureus contributes to the pathophysiology or merely reflects the abnormal environment has never been conclusively established. Molecular techniques enable a more comprehensive characterization of the changing cutaneous microbiota in skin disease and its relationship to clinical features.3,4
In this study, in the context of a clinical trial comparing topical corticosteroids (TCS) alone to TCS plus dilute bleach baths (BB) for AD treatment, we used high throughput DNA sequencing and quantitative PCR to characterize the cutaneous microbiota of children with AD and controls. Comparison of lesional and nonlesional skin with that of controls at baseline and over the course of the two regimens enabled a more comprehensive view of microbiota characteristics and their changes with disease activity. Our results show that in addition to its clinical efficacy in treating AD, TCS treatment also normalizes cutaneous microbiota compositions in AD, even without added BB.
METHODS
Patients and study design
Children of age 3 months-5 years with moderate-to-severe AD (based on modified Hanifin and Rajka criteria5) were enrolled from the pediatric dermatology clinics at the Charles C. Harris Skin and Cancer Unit (SCU) and NYU Dermatologic Associates, both at NYU Langone Medical Center, and at Bellevue Hospital Center (BHC). Control patients were enrolled from the adjacent general pediatric clinics if they were in the correct age group, and did not have any skin disease. This study was approved by the NYULMC Institutional Review Board.
Exclusion criteria
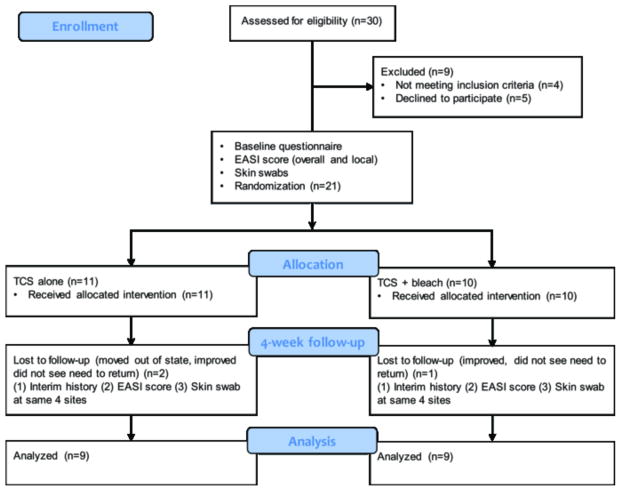
Patients with concurrent chronic inflammatory skin disorders or who were currently using or had used systemic or topical antibiotics, corticosteroids, or calcineurin inhibitors for AD in the prior two weeks were excluded from the study, as were children with overt infection. See Figure 1 for study design.6
Figure 1. Study Design.
Twenty-one subjects with AD and 14 controls were enrolled. Moderate to severe AD was defined as an investigator’s global assessment (IGA) score of 3 or 4 (0–4 scale). At baseline, a detailed questionnaire was administered, local and overall Eczema Area and Severity Index (EASI) scores were recorded and skin swabs were obtained from 4 clinical sites (3 lesional and 1 nonlesional) in AD patients and at 4 sites of AD predilection in control patients. Ten AD subjects were randomized to receive a bottle of bleach to be diluted into bath water twice weekly to achieve a 0.005% sodium hypochlorite concentration, while 11 (TCS alone group) received an identical bottle containing water and the same instructions for its use. At 4-week follow-up, parents were asked about treatment use, and local and overall EASI scores were again recorded and the 4 same sites were swabbed.
Randomization
Treatment randomization was done by two independent non-clinical staff members. Twelve sealed envelopes containing the word bleach and 12 identical sealed envelopes containing the word water were made by one person, which were then shuffled and numbered sequentially. The envelopes were opened in number order and plain white bottles were filled with the corresponding ingredient, bleach or water, and then labeled with the corresponding number.
Sampling
All AD patients had four clinical sites swabbed including the worst affected site (i.e. the lesion with the highest local EASI score), a nonlesional site, and 2 other representative lesional sites. Control patients were swabbed at the typical age-specific areas of eczema appearance. When possible, the nonlesional sample was taken from a site contralateral to the lesional site; however, when the presence of symmetric disease rendered this impossible, we selected an adjacent uninvolved area. Specimens were obtained using a sterile cotton-tipped applicator pre-moistened with 0.9% normal saline and 0.1% Tween 20, and swabbing the skin at the specified site with moderate pressure for 40 seconds, as exactly described.7 After centrifugation, the fluid was used for DNA extraction, using the MoBio Powersoil kit (Carlsbad CA) per the manufacturer’s protocol.
Quantitative PCR
TaqMan quantitative PCR (qPCR) was performed using specific primers and probes for the 16S rRNA sequences of Staphylococcus aureus, other Staphylococcus species, Corynebacterium, Proprionibacterium, Streptococcus, and universal bacteria exactly as described.8 S. aureus was amplified using the primers to target the S.aureus-specific nuc gene.
High throughput DNA Sequencing
For each sample, the V4 region of bacterial 16S rRNA genes was amplified in triplicate reactions using the universal bacterial primer set 515F/806R, as described.9,10
Statistical analysis
Comparisons of bacterial densities between cutaneous sites or between moist, dry, or sebaceous groups at different times were performed by the repeated measures ANOVA test with post hoc paired comparison. The compound symmetry correlation structure was used to account for the correlation between different samples from the same patient. Comparison of bacterial densities between treatments was performed by paired t- test. The relationship between bacterial density and EASI was analyzed with Spearman correlation. P-values <0.05 were considered to be significant and false discovery rate (FDR) was used to control for multiple comparisons. See Supplementary Methods for statistical analysis of sequencing data.
RESULTS
Patients
There were no significant differences in sex, age, or race between the 35 enrolled AD patients or the 29 subjects who completed the study and controls (Table S1), nor in the clinical severity scores between the subjects randomized to the two treatment groups. Three subjects with AD (two in placebo group, one in bleach group) and three control group subjects did not receive follow-up assessment due to their moving out of state or not returning for the examination. When contacted, parents of all three of these AD patients reported improvement of their child’s eczema. Among patients who completed the study, the mean age in the TCS alone group was lower than in the TCS + bleach and control groups (Table S1). As expected, AD patients reported a family history of AD more often than controls, a significant difference for the TCS + bleach group (p=0.002), but not for the TCS alone group, after adjustment for multiple comparisons. There were no significant differences among the AD groups and controls in proportions of subjects who were born via vaginal delivery or bathed less than daily, or in the mean time from the last bath or topical cream application.
Clinical outcome
All 18 AD subjects who completed the trial had significant improvement in clinical scores; both treatment groups had significant improvements in IGA, total EASI and local EASI scores over time (Figure S1A–C), with no significant differences between the groups. Only two subjects in our cohort were of school age (>48 months old). We have analyzed the change in EASI with respect to age, controlling for treatment, mode of delivery and skin site, using a multiple linear regression model. The coefficient for age was not significant (p>0.05). Therefore, we conclude that the age of the subject did not have an effect on treatment outcome.
Baseline bacterial densities
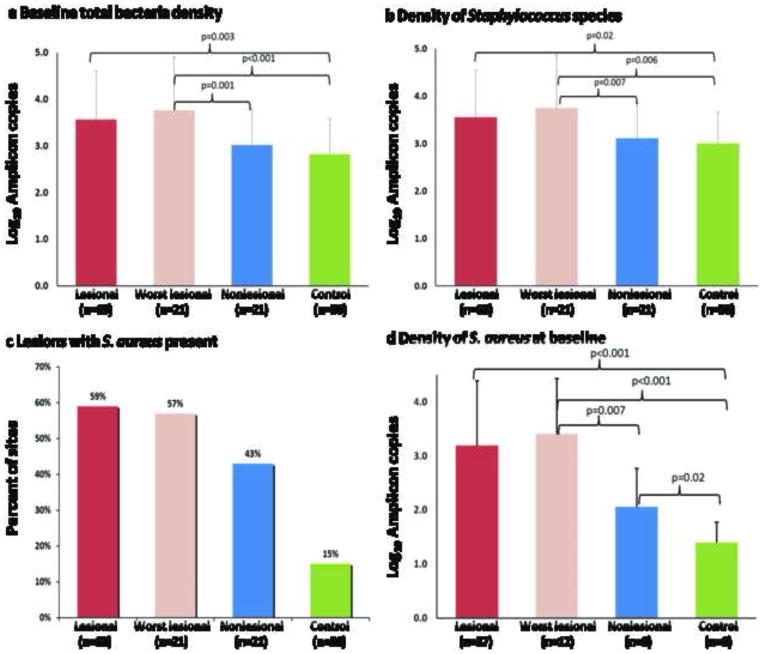
From the 21 AD patients, we sampled 63 lesional and 21 nonlesional sites. As expected, at baseline, lesional skin had higher total bacterial concentrations by qPCR than nonlesional or control skin, which did not differ from one another. The same trends were observed for Staphylococcus species (Figure 2A–D). In contrast, there were no overall differences in baseline densities of Streptococcus, Corynebacterium, or Proprionibacterium species in control versus lesional or nonlesional skin. At the worst affected sites, Corynebacterium and Proprionibacterium species densities were significantly lower than for control skin (data not shown).
Figure 2. Bacterial densities at baseline determined by specific qPCR assays.
Mean densities of all sites swabbed from all enrolled subjects at baseline are presented here. A. Baseline, lesional skin had higher total bacterial concentrations than nonlesional or control skin, which did not differ from one another. The mean total bacterial density at the worst affected site was 4 log10/ sample, which represents ~5-fold greater density than on nonlesional skin in the same subject, and ~10-fold greater density than on control skin. B. The same trends were observed for Staphylococcus. C. At baseline, S. aureus was detected in a higher proportion of the worst affected lesional sites than in the control sites. D. The mean S. aureus density in lesions in which it was detected was significantly higher in the worst affected sites than on control or nonlesional sites. Symbols: T1, baseline; T2, follow-up visit.
As expected, densities of total bacteria and predominant skin genera varied by microenvironment (moist, dry and sebaceous) (Figure S2A–E). At baseline for dry skin sites, known to harbor the greatest diversity11, total bacterial counts on lesional skin were significantly higher than for controls. At sebaceous sites, known to be the least diverse, S. aureus density on lesional skin was significantly higher than control skin although there were no significant differences in genus Staphylococcus. Corynebacterium was greater on control dry sites than dry sites from lesional AD skin (Figure S2A–E). Bacterial densities were similar for control and nonlesional AD skin at dry and sebaceous sites.
Correlation of EASI clinical score with bacterial densities
Mean lesional total bacterial density and mean lesional S. aureus density each were moderately correlated with overall EASI scores; (Figure S3A–B). Patients in whom S. aureus was not detected tended to have lower total EASI scores.
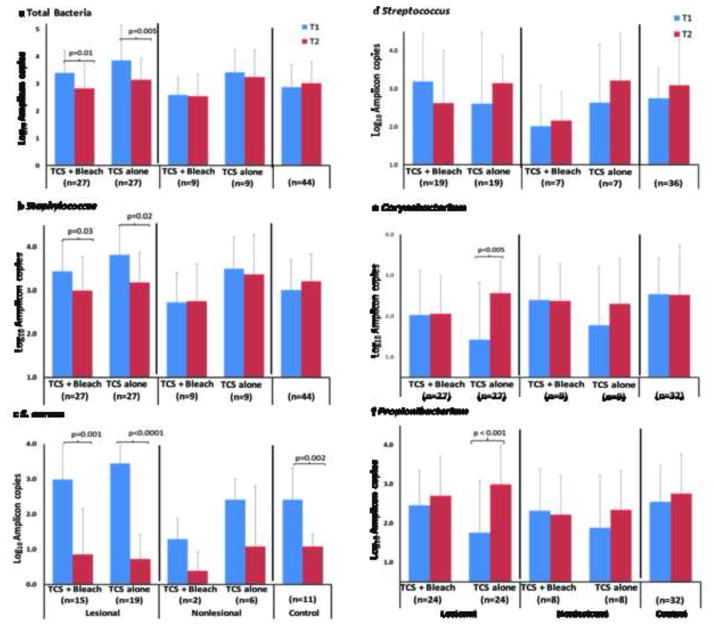
Effect of treatment on bacterial densities
The effect of TCS + bleach bath versus TCS alone on bacterial densities is shown in Figure 3A–F.
Figure 3. Effect of TCS + bleach versus TCS alone on bacterial densities.
Densities determined by specific qPCR assays. A. Both treatments (TCS + bleach and TCS alone) significantly lowered total bacterial densities on lesional sites, but not on nonlesional sites or control skin. B. Total bacterial and Staphylococcus densities on lesional skin became similar to control skin. On nonlesional skin, Staphylococcus densities remained stable. C. The S. aureus density significantly decreased in both treatment groups on lesional sites and unexpectedly in controls and trended down on nonlesional skin. D. Streptococcus species did not change significantly with treatment. E &F. Corynebacterium and Propionibacterium densities did not change on lesional or nonlesional skin after TCS + bleach treatment but increased on lesional skin in those who received TCS alone. Symbols: T1, baseline; T2, follow-up visit.
Microbiota status
In total, 256 samples were analyzed, yielding 1,246,175 sequences in total [median 4,297 sequences per sample (IQR = 2,700 – 6,163)]. To ensure valid comparisons, we examined the 249 samples with > 1,000 sequences available.
Alpha diversity
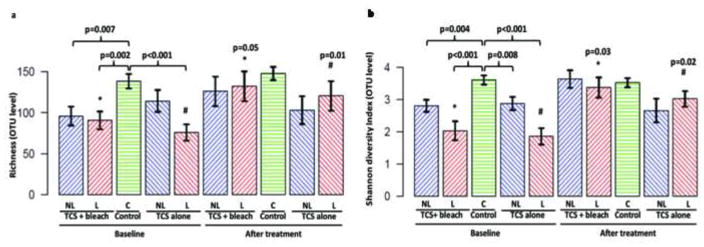
The mean Shannon diversity index (SDI) of microbial communities at all lesional sites was inversely correlated with overall EASI score at baseline (Figure S3C). Both treatments resulted in decreased overall EASI scores, and higher diversity scores (Figure S3D). Using linear regression, the follow-up EASI scores by treatment group were not significantly associated (p>0.05) with Shannon and richness diversities; thus correlation of disease severity with diversity was equally lost with both successful treatments.
Significant increases in bacterial community richness and SDI after both treatments are shown in Figure 4. Each cutaneous microenvironment showed similar trends (Figure S4).
Figure 4. Diversity scores for control, lesional, and nonlesional sites.
At baseline, bacterial community richness and SDI at both lesional and nonlesional sites in both treatment groups were lower than control skin, to similar extents. A. Following treatment, the mean OTU numbers (representing community richness) increased at lesional sites, becoming similar to control skin in both treatment groups. B. After treatment, there were no changes in the mean Shannon diversity index in control or nonlesional skin but diversity for lesional skin for both treatment groups normalized, resembling control skin. Symbols: L, lesion; NL, uninvolved site; C, control site.
Community structure
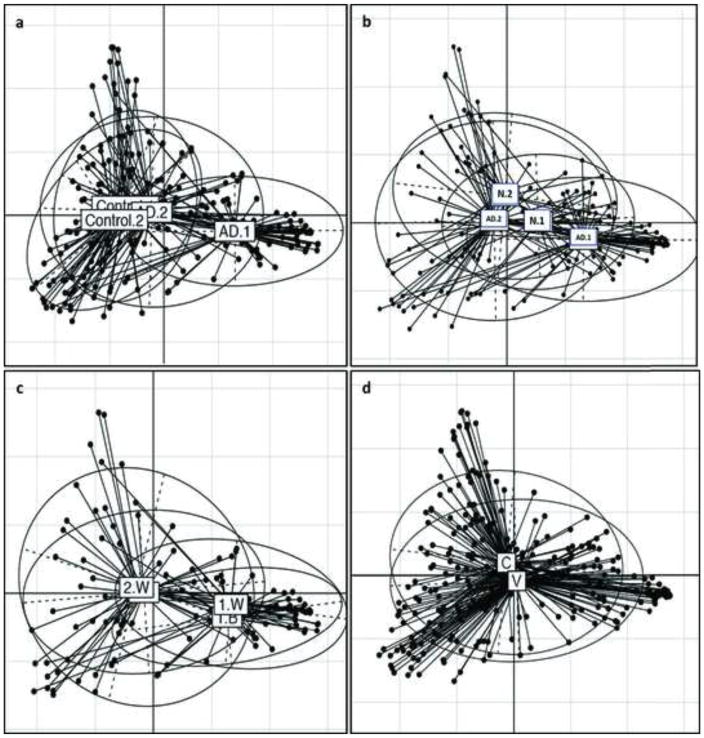
Community structure (β-diversity) assessed by Unifrac analysis after controlling for method of delivery (vaginal or Caesarean-section) is shown in Figures 5A–D.
Figure 5. Principal Coordinates Analysis (PCoA) of microbial community composition.
A. Compositions of bacterial communities on lesional AD skin were significantly different than control sites at baseline. After either treatment, communities on lesional AD skin approached that of control skin but remained significantly different. The bacterial communities on control skin was not different at the two sampling timepoints. B. At baseline, lesional and nonlesional communities differed, but not significantly. After treatment, bacterial communities on lesional AD skin significantly differed from baseline, becoming similar to nonlesional skin. The nonlesional sites did not significantly change after treatment. C. The bacterial communities on lesional sites in the two treatment groups were similar to each other at baseline and after treatment. D. There was no significant difference in the bacterial communities of vaginally-delivered versus subjects delivered by C-section at baseline. Symbols: AD: Lesional; N: nonlesional; B: TCS + bleach; W: TCS alone.
Taxa abundance and changes in microbiome composition
Taxonomic normalization occurred on the lesional and nonlesional microbiota after treatment in both groups. The relative abundance of major phyla and major genera in control and AD lesional and nonlesional sites at baseline and after treatment is shown in Figures 6 and S5, respectively.
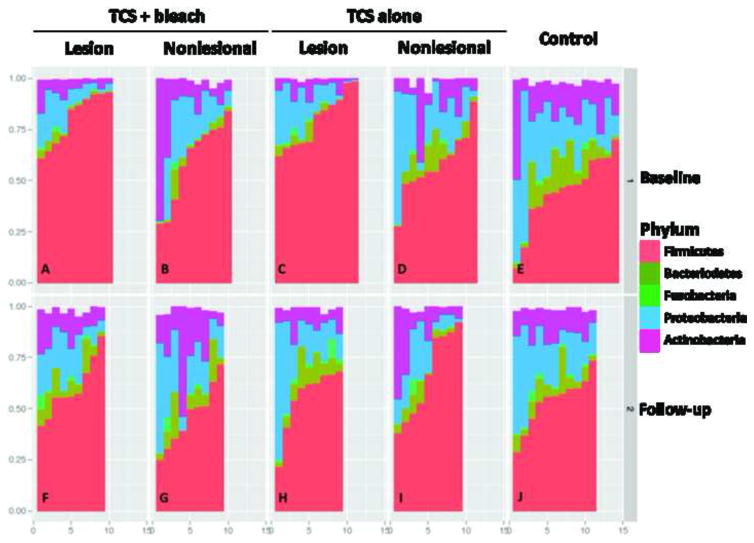
Figure 6. Relative abundance of major phyla in control and AD lesional and nonlesional sites at baseline and after treatment.
Analysis performed at sequence depth > 4,800 reads for each of the 249 specimens. At baseline (Panels A–E), all sites showed a mixture of five major phyla but for AD lesional skin, Firmicutes dominated with relative absence of Bacteriodetes (Panels A,C). Nonlesional skin showed intermediate patterns (Panels B, D). After the treatment period (Panels F–J), controls were largely unchanged, as expected (Panel J), but all AD sites normalized (Panel F) with decreased Firmicutes, and Bacteroidetes reappearing. Control subjects: E,J; TCS + bleach treatment group: lesional sites: A,J; nonlesional sites; B,G; TCS alone group: lesional sites: C,H; nonlesional sites: D,I.
In most controls at baseline, 13 major genera comprised < 50% of all reads (Figure S5), with Staphylococci generally < 25%, indicating high diversity in the lower abundance genera. In contrast, the median representation of Staphylococci in lesional skin was 60–70% (Panels A, C), and nonlesional skin showed intermediate patterns (Panels B, D). At the 4-week follow up, there was little change in controls (Panel J), but both lesional and the nonlesional AD samples normalized (Panels F–I). On nonlesional skin, bleach baths lead to a greater suppression of Staphylococcus species compared to the TCS group (Panel G).
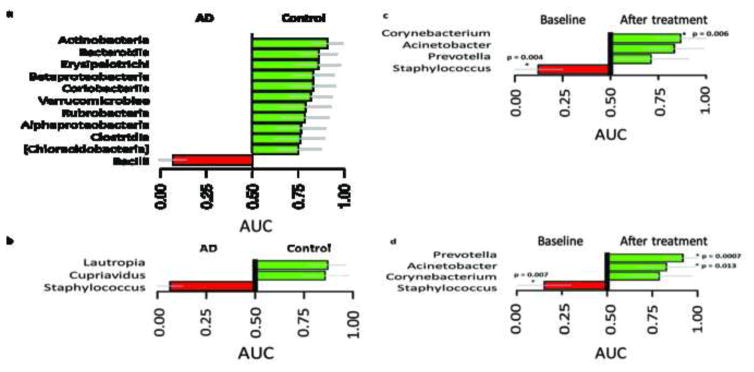
Area under the curve (AUCS) plots showing differentiating taxa and differentiating genera with ≥≥1% abundance can be seen in Figure 7A–B and 7C–D respectively.
Figure 7. Area under the curve (AUCS) plots showing differentiating taxa (A & B) and differentiating genera with ≥≥ 1% abundance (C & D).
A. Baseline AD lesional sites versus control skin at the class level. At baseline, Bacilli were significantly over-represented on lesional AD skin compared to control skin (AUC = 0.01; p<0.001), whereas Actinobacteria and several other taxa were significantly over-represented in controls. B. Baseline AD lesional sites versus control skin at the genus level. At the genus level, Staphylococcus was significantly overrepresented at baseline for lesional compared to control skin, where Lautropia and Cupriavidus were significantly over-represented in control. C. TCS + bleach treated group at baseline and after treatment. On lesional skin, Staphylococcus was significantly over-represented at baseline and after treatment, Staphylococcus was replaced by Corynebacterium (AUC = 0.87, p=0.006). D. TCS alone group at baseline and after treatment. On lesional skin Staphylococcus was significantly over-represented at baseline and after treatment, Prevotella (AUC = 0.92, p=0.007) and Acinetobacter (AUC = 0.83, p=0.01) were significantly enriched. Significance defined as p<0.05, based on Mann-Whitney rank sum test and FDR adjusted.
DISCUSSION
High throughput sequencing of the conserved 16S rRNA gene sequences has allowed in-depth profiling of the cutaneous microbiota and is advancing our knowledge of microbial roles in disease.12,13,14,15 Studies of inflammatory diseases including psoriasis and atopic dermatitis indicate associations with cutaneous microbiota alterations.3,16,17,18 Whether the altered microbial composition has a pathogenic role or reflects selection of particular taxa due to epidermal and immune abnormalities inherent to the inflammatory condition remains to be fully determined.
In this study, genus-specific quantitative PCR confirmed the findings of culture-based studies showing that patients with atopic dermatitis have increased colonization with S.aureus on both lesional and nonlesional skin.19,20,21,22,23 In addition, S. aureus density correlated with disease severity (by EASI score), confirming earlier studies.24,25
In a study of the cutaneous microbiome using near-full-length sequencing of 16S rRNA genes, Kong et al. show that AD lesional skin during disease flares had increased abundance of S. aureus and S. epidermidis, but decreased Streptococcus, Corynebacterium, and Propionibacterium; disease severity and cutaneous bacterial diversity were inversely associated. The patients they studied were given varying combinations of treatments (TCS, BB, and topical and systemic antibiotics), yet microbial diversity was restored to baseline levels, resulting from re-expansion of taxa already present at baseline. In that study, “baseline” referred to a stable disease state and was often obtained several months after the flare and post-flare time points. Since multiple treatments, including anti-bacterial and anti-inflammatory modalities, were used, it is not clear which treatment led to the normalization of diversity.
The current study was a prospective controlled clinical trial in which all AD patients had no active treatments in the preceding 2 weeks; all subjects were prescribed fluticasone cream and then randomized to either bleach baths or placebo. Sequencing the V4 region of 16S rRNA allowed for genus-level characterization and greater (median ~4300) sequencing depth, that can be compared with the many other current studies using V4 sequencing.26,27,28 Patients in the current study were 3 months to 5 years of age, representing a more homogeneous AD population than in prior studies and the age range most closely associated with disease emergence. The majority of patients in the current study were less than 2 years old; and thus the current study is the first to use molecular techniques to characterize the cutaneous microbiota in well infants and young children, and those with AD.
We show that overall microbial diversity was inversely correlated with disease severity, as low diversity was associated with relatively high Staphylococcus relative and absolute abundances. Nonlesional skin also exhibited decreased diversity relative to controls, suggesting an altered cutaneous microbiota beyond clinically affected skin in AD patients. While culture-based studies have shown increased rates of Staphylococcus colonization on nonlesional skin,29,30 molecular characterization of the nonlesional cutaneous microbiome and how it differs from control skin using both qPCR and 16Ss sequencing had not been reported.
In addition, we show that baseline bacterial communities on both lesional and nonlesional AD skin differed in composition from controls. After the immunomodulating TCS treatment with or without BB, lesional bacterial community compositions resembled nonlesional skin but remained different from controls, pointing to primary microbiota abnormalities in AD.
Several studies have demonstrated that nonlesional AD skin is different from normal skin with respect to terminal differentiation and immunity.31,32 Atopic skin, both lesional and nonlesional, is associated with increased transepidermal water loss, increased permeability to topical agents,33 and reduced antimicrobial peptides.34 Filaggrin (FLG) gene mutations, which lead to higher pH and decreased moisture across cutaneous surfaces,35 are associated with increased AD severity and duration plus predisposition to cutaneous infection, and increased S. aureus colonization.36,37 Filaggrin expression is also decreased in nonlesional AD skin. Similarly, cutaneous bacterial compositions differ from control in immunodeficient patients,38 and the skin of ADAM17-deficient mice.39 Our observations suggest that in addition to alterations in cutaneous immunity, nonlesional skin demonstrates a unique microbial community composition.
TCS treatment with or without BB led to similar significant clinical improvements, associated with restoration of microbial diversity and decreased numbers of total bacteria, S. aureus, and other Staphylococcus species on lesional skin. Our findings are consistent with an updated Cochrane review concluding that although anti-staphylococcal interventions may reduce S. aureus density in AD patients, there was no added clinical benefit compared to anti-inflammatory interventions.40 A randomized study in pediatric AD patients who first received two weeks of oral antibiotic for skin infections showed greater EASI score reduction with twice-weekly BB and intranasal mupirocin for 3 months than with placebo; clinical benefit was observed despite continued cutaneous S. aureus presence.41 Another study of pediatric and adult AD patients showed that twice-weekly BB led to a lower 2-month EASI score than TCS and emollient use alone; the BB diminished but did not eradicate S. aureus.42 Our four-week follow-up time may not have been sufficient to observe BB benefits, and the marked EASI score reduction in both groups left little room for additional benefit. It is possible that anti-inflammatory as well as antimicrobial effects of sodium hypochlorite contribute to BB-related improvement of AD.43
We observed increased overall cutaneous microbial diversity with SDI returning to the same levels as control skin after both treatments, however the composition of the bacterial communities after treatment resembles nonlesional skin and is distinct from control skin. Our results suggest that TCS suffices to normalize the cutaneous microbial communities on lesional skin. We cannot rule out a contribution from excipients present in the fluticasone proprionate cream. After both treatments, S. aureus was replaced by Corynebacterium, Prevotella, and Acinetobacter (Figure 7). Corynebacteria, one of the most abundant genera on healthy human skin may be inversely related to S. aureus from the nares.44 Although not major cutaneous taxa, Acinetobacter and Prevotella may be normal bacterial biota in moist intertriginous areas,45,46 and Prevotella species, have been found on the skin of vaginally-delivered newborns.47
While sequencing the V4 region allowed us to detect differential effects of bleach (Figure 7 & S5), the 4-week time period and/or the twice weekly baths may not have been sufficient for the added treatment to have a meaningful impact on the cutaneous microbiome. In addition, children in this study were younger on average than prior studies showing clinical effects of bleach. More detailed sequencing (sequencing of V1-3 or whole 16S rRNA gene) in future studies may allow more detailed characterization of the distinguishing genera in bleach bath treated patients.
Deciphering complex cutaneous host-microbe and microbe-microbe interactions and their roles in skin disease and innate immunity is a daunting task. Certain commensals, such as S. epidermidis and P. acnes, may inhibit S. aureus,48,49,50,51 and their reemergence may help restore balanced communities. Recent evidence suggests that nonpathogenic bacteria alleviates AD related skin inflammation.52 Conversely, AD-related perturbations in S. aureus populations may be secondary to the disease process, largely a function of the disrupted barrier function, rather than representing primary etiologic events.53 Marked reductions in S. aureus levels in atopic dermatitis have been observed in subjects treated with topical corticosteroids or tacrolimus ointment.54,55 Our study adds to the evidence that decreasing inflammation and improving barrier function may suffice to reduce S. aureus and normalize AD bacterial compositions.3,56 Dilute sodium hypochlorite (bleach) baths have been recommended to patients with atopic dermatitis by dermatologists for decades. Few studies have rigorously examined the utility of this measure. Our study did not find any microbiologic benefits beyond fluticasone proprionate cream in the acute treatment setting. Further studies are needed to definitively establish the value of dilute bleach baths in the long-term maintenance of AD.
Supplementary Material
Acknowledgments
This study was approved by the NYULMC Institutional Review Board (protocol #10-01622).
Funding sources. ME Gonzalez was a recipient of Pediatric Dermatology Fellowship Award from the Dermatology Foundation and JV Schaffer received a Medical Dermatology Career Development Award from the Dermatology Foundation. ME Gonzalez was supported in part by a gift from the Bohnert Foundation. This work has utilized computing resources at the High Performance Computing Facility of the Center for Health Informatics and Bioinformatics at the NYU Langone Medical Center. Supported in part by UH2 AR057506 and NYU CTSA grant UL1 TR000038 from the National Institutes of Health, and by the Diane Belfer Program in Human Microbial Ecology, and C&D Research Fund.
Footnotes
Conflict of interest statement.
The authors of this manuscript do not have any relevant conflicts of interest to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eichenfield LF, Ellis CN, Mancini AJ, et al. Atopic dermatitis: epidemiology and pathogenesis update. Semin Cutan Med Surg. 2012;31(3 Suppl):S3–5. doi: 10.1016/j.sder.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Biedermann T. Dissecting the Role of Infections in Atopic Dermatitis. Acta Derm Venereol. 2006;86:99–109. doi: 10.2340/00015555-0047. [DOI] [PubMed] [Google Scholar]
- 3.Kong HH, Segre JA. Skin microbiome: looking back to move forward. J Invest Dermatol. 2012;132:933–39. doi: 10.1038/jid.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouslimani A, Porto C, Rath CM, et al. Molecular cartography of the human skin surface in 3D. Proc Natl Acad Sci. 2015;112(17):E2120–9. doi: 10.1073/pnas.1424409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichenfield LF, Hanifin JM, Luger TA, et al. Consensus conference on pediatric atopic dermatitis. J Am Acad Dermatol. 2003;49:1088–1095. doi: 10.1016/s0190-9622(03)02539-8. [DOI] [PubMed] [Google Scholar]
- 6.Schmitt J, Langan S, Deckert S, et al. Assessment of clinical signs of atopic dermatitis: a systematic review and recommendation. J Allergy Clin Immunol. 2013;132(6):1337–47. doi: 10.1016/j.jaci.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Gao Z, Perez IP, Chen Y, et al. Quantitation of major human cutaneous bacterial and fungal populations. J Clin Microbiol. 2010;48:3575–81. doi: 10.1128/JCM.00597-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redel H, Gao Z, Li H, et al. Quantitation and composition of cutaneous microbiota in diabetic and nondiabetic men. J Infect Dis. 2013;207:1105–14. doi: 10.1093/infdis/jit005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walters WA, Caporaso JG, Lauber CL, et al. Primer-Prospector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics. 2011;27:1159–1161. doi: 10.1093/bioinformatics/btr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high- throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME Journal. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–92. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–707. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Findley K, Oh J, Yang J, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conlon S, Kong HH, Segre JA. Species-level analysis of DNA sequence data from the NIH Human Microbiome Project. PLoS One. 2012;7:e47075. doi: 10.1371/journal.pone.0047075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Z, Tseng CH, Strober BE, et al. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS One. 2008;3:e2719. doi: 10.1371/journal.pone.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahlén A, Engstrand L, Baker BS, et al. Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Arch Dermatol Res. 2012;304:15–22. doi: 10.1007/s00403-011-1189-x. [DOI] [PubMed] [Google Scholar]
- 18.Alekseyenko AV, Perez-Perez GI, De Souza A, et al. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome. 2013;23:31. doi: 10.1186/2049-2618-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leyden JJ, Marples RR, Kligman AM. Staphylococcus aureus in the lesions of atopic dermatitis. Br J Dermatol. 1974;90:525–30. doi: 10.1111/j.1365-2133.1974.tb06447.x. [DOI] [PubMed] [Google Scholar]
- 20.Higaki S, Morohashi M, Yamagishi T, et al. Comparative study of staphylococci from the skin of atopic dermatitis patients and from healthy subjects. Int J Dermatol. 1999;38:265–269. doi: 10.1046/j.1365-4362.1999.00686.x. [DOI] [PubMed] [Google Scholar]
- 21.Gong JQ, Lin L, Lin T, et al. Skin colonization by Staphylococcus aureus in patients with eczema and atopic dermatitis and relevant combined topical therapy: a double-blind multicentre randomized controlled trial. Br J Dermatol. 2006;155:680–87. doi: 10.1111/j.1365-2133.2006.07410.x. [DOI] [PubMed] [Google Scholar]
- 22.Breuer K, Haussler S, Kapp A, et al. Staphylococcus aureus: colonizing features and influence of an antibacterial treatment in adults with atopic dermatitis. Br J Dermatol. 2002;147:55–61. doi: 10.1046/j.1365-2133.2002.04872.x. [DOI] [PubMed] [Google Scholar]
- 23.Park HY, Kim CR, Huh IS, et al. Staphylococcus aureus Colonization in Acute and Chronic Skin Lesions of Patients with Atopic Dermatitis. Ann Dermatol. 2013;25(4):410–6. doi: 10.5021/ad.2013.25.4.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nilsson EJ, Henning CG, Magnusson J. Topical corticosteroids and Staphylococcus aureus in atopic dermatitis. J Am Acad Dermatol. 1992;27:29–34. doi: 10.1016/0190-9622(92)70151-5. [DOI] [PubMed] [Google Scholar]
- 25.Guzik TJ, Bzowska M, Kasprowicz A, et al. Persistent skin colonization with Staphylococcus aureus in atopic dermatitis: relationship to clinical and immunological parameters. Clin Exp Allergy. 2005;35(4):448–55. doi: 10.1111/j.1365-2222.2005.02210.x. [DOI] [PubMed] [Google Scholar]
- 26.Clemente JC, Pehrsson EC, Blaser MJ, et al. The microbiome of uncontacted Amerindians. Science Advances. 2015;1(3):1–12. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton JM, Gao Z, Sullivan M, et al. The cutaneous microbiome in out-patients presenting acutely with skin abscesses. J Infect Dis. 2015;211(12):1895–1904. doi: 10.1093/infdis/jiv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meadow JF, Bateman AC, Herkert KM, et al. Significant changes in the skin microbiome mediated by the sport of roller derby. Peer J. 2013;1:e53–70. doi: 10.7717/peerj.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsui K, Nishikawa A, Suto H, et al. Comparative study of Staphylococcus aureus isolated from lesional and nonlesional skin of atopic dermatitis patients. Microbiol Immunol. 2000;44(11):945–7. doi: 10.1111/j.1348-0421.2000.tb02587.x. [DOI] [PubMed] [Google Scholar]
- 30.Kedzierska A, Kapińska-Mrowiecka M, Czubak-Macugowska M, et al. Susceptibility testing and resistance phenotype detection in Staphylococcus aureus strains isolated from patients with atopic dermatitis, with apparent and recurrent skin colonization. Br J Dermatol. 2008;159(6):1290–9. doi: 10.1111/j.1365-2133.2008.08817.x. [DOI] [PubMed] [Google Scholar]
- 31.Suarez-Farinas M, Tintle S, Shemer A, et al. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J Allergy Clin Immunol. 2011;127(4):954–64. doi: 10.1016/j.jaci.2010.12.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee HJ, Lee SH. Epidermal Permeability Barrier Defects and Barrier Repair Therapy in Atopic Dermatitis. Allergy Asthma Immunol Res. 2014;6(4):276–87. doi: 10.4168/aair.2014.6.4.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Benedetto A, Slifka MK, Rafaels NM, et al. Reductions in claudin-1 may enhance susceptibility to herpes simplex virus 1 infections in atopic dermatitis. J Allergy Clin Immunol. 2011;128(6):1216–24. doi: 10.1016/j.jaci.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ong PY, Ohtake T, Brandt C, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med. 2002;347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 35.Nakamizo S, Egawa G, Honda T, et al. Commensal bacteria and cutaneous immunity. Semin Immunopathol. 2015;37(1):73–80. doi: 10.1007/s00281-014-0452-6. [DOI] [PubMed] [Google Scholar]
- 36.Van Drongelen VV, Haisma EM, Out-Luiting JJ, et al. Reduced filaggrin expression is accompanied by increased Staphylococcus aureus colonization of epidermal skin models. Clin Exp Allergy. 2014;44:1515–24. doi: 10.1111/cea.12443. [DOI] [PubMed] [Google Scholar]
- 37.Gao PS, Rafaels NM, Hand T, et al. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J Allergy Clin Immunol. 2009;124(3):507–13. doi: 10.1016/j.jaci.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh J, Freeman AF, et al. NISC Comparative Sequencing Program. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res. 2013;23:2103–14. doi: 10.1101/gr.159467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi T, Glatz M, Horiuchi K, et al. Dysbiosis and Staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity. 2015;42:756–66. doi: 10.1016/j.immuni.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bath-Hextall FJ, Birnie AJ, Ravenscroft JC, et al. Interventions to reduce Staphylococcus aureus in the management of atopic eczema: an updated Cochrane review. Br J Dermatol. 2010;163:12–26. doi: 10.1111/j.1365-2133.2010.09743.x. [DOI] [PubMed] [Google Scholar]
- 41.Huang JT, Abrams M, Tlougan B, et al. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics. 2009;123:e808–14. doi: 10.1542/peds.2008-2217. [DOI] [PubMed] [Google Scholar]
- 42.Wong SM, Ng TG, Baba RJ, et al. Efficacy and safety of sodium hypochlorite (bleach) baths in patients with moderate to severe atopic dermatitis in Malaysia. Dermatol. 2013;40:874–80. doi: 10.1111/1346-8138.12265. [DOI] [PubMed] [Google Scholar]
- 43.Leung TH, Zhang LF, Wang J, et al. Topical hypochlorite ameliorates NF-kB mediated skin diseases in mice. J Clin Invest. 2013;123(12):5361–70. doi: 10.1172/JCI70895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uehara Y, Nakama H, Agematsu K, et al. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J Hosp Infect. 2000;44:127–33. doi: 10.1053/jhin.1999.0680. [DOI] [PubMed] [Google Scholar]
- 45.Seifert H, Dijkshoorn L, Gerner-Smidt P, et al. Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods. J Clin Microbiol. 1997;35:2819–25. doi: 10.1128/jcm.35.11.2819-2825.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiller K, Selkin BA, Murakawa GJ. Skin microflora and bacterial infections of the skin. J Investig Dermatol Symp Proc. 2001;6:170–74. doi: 10.1046/j.0022-202x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 47.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci. 2010;107:11971–75. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwase T, Uehara Y, Shinji H, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2020;465:346–49. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 49.Lai Y, Di Nardo A, Nakatsuji T, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15(12):1377–82. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braff MH, Bardan A, Nizet V, et al. Cutaneous defense mechanisms by antimicrobial peptides. J Invest Dermatol. 2005;125(1):9–13. doi: 10.1111/j.0022-202X.2004.23587.x. [DOI] [PubMed] [Google Scholar]
- 51.Shu M, Wang Y, Yu J, et al. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS One. 2013;8:e55380. doi: 10.1371/journal.pone.0055380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volz T, Skabytska Y, Guenova E, et al. Nonpathogenic bacteria alleviating atopic dermatitis inflammation induce IL–10 Producing Dendritic Cells and Regulatory Tr1 Cells. J Invest Dermatol. 2014;134:96–104. doi: 10.1038/jid.2013.291. [DOI] [PubMed] [Google Scholar]
- 53.Findley K, Grice EA. The skin microbiome: a focus on pathogens and their association with skin disease. PLoS Pathog. 2014;10(10):e1004436. doi: 10.1371/journal.ppat.1004436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stadler JF, Fleury M, Sourisse M, et al. Local steroid therapy and bacterial skin flora in atopic dermatitis. Br J Dermatol. 1994;131:536– 540. doi: 10.1111/j.1365-2133.1994.tb08556.x. [DOI] [PubMed] [Google Scholar]
- 55.Hung SH, Lin YT, Chu CY, et al. Staphylococcus colonization in atopic dermatitis treated with fluticasone or tacrolimus with or without antibiotics. Ann Allergy Asthma Immunol. 2007;98(1):51–6. doi: 10.1016/S1081-1206(10)60859-9. [DOI] [PubMed] [Google Scholar]
- 56.Flores GE, Seite S, Henley JB, et al. Microbiome of Affected and Unaffected Skin of Patients With Atopic Dermatitis Before and After Emollient Treatment. J Drugs Dermatol. 2014;13:1365–72. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.