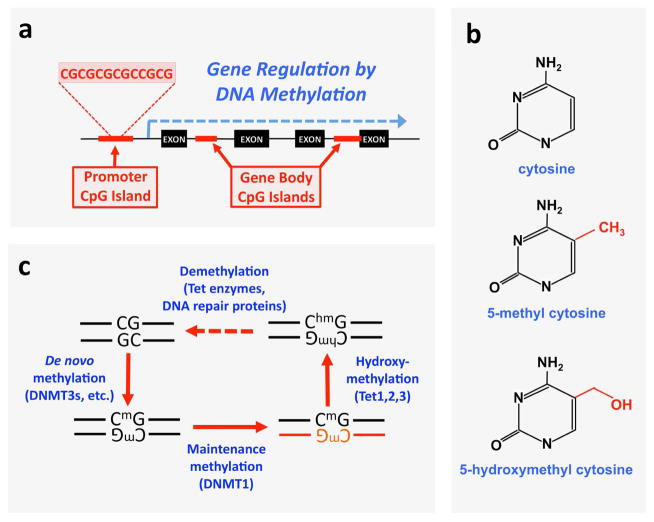
Figure 1. DNA methylation and demethylation processes.
a, Schematic showing the relative positions of the CpG islands associated with a gene locus. Promoter CpG islands are found in upstream regulatory regions, and gene body CpG islands are present anywhere between the start and stop codon. Promoter CpG islands are involved in gene silencing, while recent evidence suggests that gene body CpG islands promote gene activation, b, Chemical structure of cytosine, 5-methyl cytosine, and 5-hydroxymethyl cytosine. c, Methylation and demethylation is driven by distinct sets of enzymes in vivo. Newly established methyl “marks” are added by de novo methyltransferases, including the Dnmt3 family. The maintenance methyltransferase Dnmt1 ensures persistence of these marks in replicating DNA by copying the methyl marks onto to newly synthesized strands (red) during mitosis. Tet proteins hydroxylate methyl groups on cytosine, with subsequent removal of the hydroxymethyl tag by Tet and other DNA repair enzymes leading back to unmethylated DNA.