Abstract
PSM-mec is a secreted virulence factor that belongs to the phenol-soluble modulin (PSM) family of amphipathic, alpha-helical peptide toxins produced by Staphylococcus species. All known PSMs are core genome-encoded with the exception of PSM-mec, whose gene is found in specific sub-types of SCCmec methicillin resistance mobile genetic elements present in methicillin-resistant Staphylococcus aureus and coagulase-negative staphylococci. In addition to the cytolytic translational product, PSM-mec, the psm-mec locus encodes a regulatory RNA. In S. aureus, the psm-mec locus influences cytolytic capacity, methicillin resistance, biofilm formation, cell spreading, and the expression of other virulence factors, such as other PSMs, which results in a significant impact on immune evasion and disease. However, these effects are highly strain-dependent, which is possibly due to differences in PSM-mec peptide vs. psm-mec RNA-controlled effects. Here, we summarize the functional properties of PSM-mec and the psm-mec RNA molecule and their roles in staphylococcal pathogenesis and physiology.
Keywords: Staphylococcus aureus, Staphylococcus epidermidis, phenol-soluble modulin, PSM-mec, regulatory RNA, SCCmec, virulence
Introduction
Staphylococcus aureus is a serious human pathogen responsible for a multitude of human diseases, which range from acute skin and soft tissue infections to more severe illnesses such as catheter-associated bacteremia, necrotizing pneumonia, and osteomyelitis. Since the emergence of methicillin-resistant S. aureus (MRSA) in the early 1960s, S. aureus has continued to cause significant morbidity, mortality, and a considerable financial burden for public health systems. For S. aureus and its closely related cousin, Staphylococcus epidermidis, the acquisition of antibiotic resistance, coupled with the ability to attach to surfaces and form sticky, multicellular agglomerations (biofilms; Costerton, 1999) on indwelling catheters, are two defining hallmarks that account for the ongoing difficulty in treating immuno-compromised individuals (Otto, 2008) with infections caused by Hospital-Associated MRSA [HA-MRSA] (Otto, 2012) and S. epidermidis (Otto, 2009).
Genetic mutations and/or the acquisition of mobile genetic elements (MGEs) that harbor drug resistance genes are two common mechanisms that are responsible for antibiotic resistance in microorganisms. Arguably the most important staphylococcal MGE is Staphylococcus cassette chromosome mec (SCCmec), which codes for methicillin resistance. Recently, a unique locus was discovered in specific types of SSCmec elements in S. aureus and S. epidermidis, which comprises a small regulatory (sr)RNA and an embedded gene encoding a cytolytic peptide called PSM-mec (Queck et al., 2009). PSM-mec belongs to the phenol-soluble modulin (PSM) family of amphipathic, alpha-helical peptide toxins produced by Staphylococcus species (Wang et al., 2007, 2011; Diep and Otto, 2008; Otto, 2014; Cheung et al., 2014a), which collectively play an important role as virulence determinants in many facets of S. aureus and S. epidermidis pathogenesis (Wang et al., 2011; Periasamy et al., 2012; Cassat et al., 2013; Cheung et al., 2014a). Here, we will discuss the current knowledge on the PSM-mec and the psm-mec srRNA molecule, with a focus on their roles in S. aureus pathogenesis.
The distribution of the psm-mec locus in SCCmec
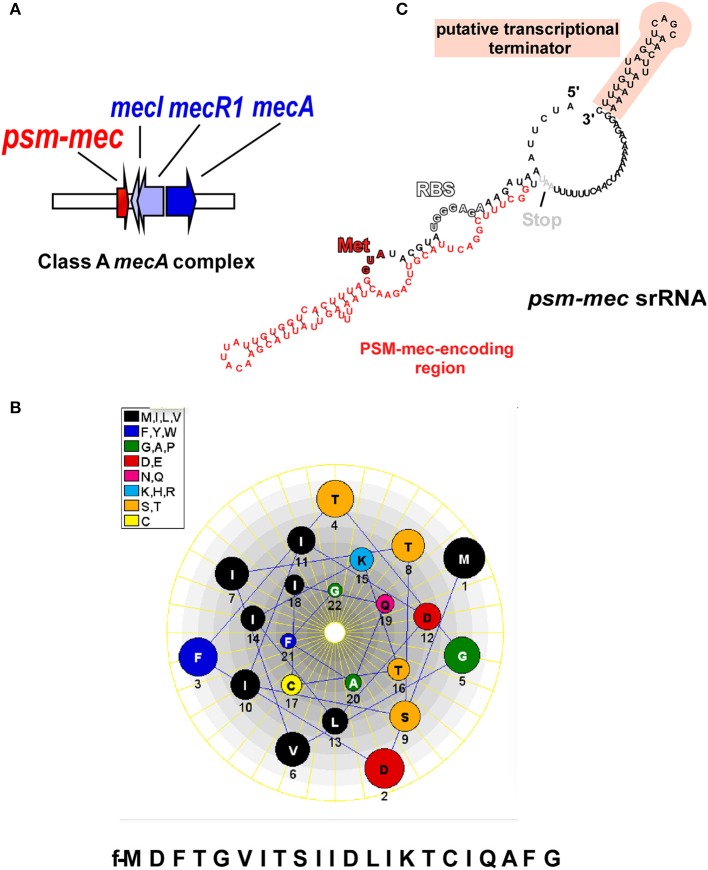
SCCmec elements, which range in size from 21 to 67 kb, comprise as characteristic components, the mecA gene that codes for a penicillin-binding protein, a ccr gene complex for site-specific recombination, and flanking repeat sequences for integration into the genome (International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements, 2009). SCCmec elements are classified into 11 different “types,” as defined by the presence of different ccr and mec resistance genes (International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements, 2009; Li et al., 2011; Shore et al., 2011) and further divided into “subtypes” based on the presence of other resistance genes and transposons in the non-essential J regions (Ito et al., 2003). The mec gene complex has four core elements; IS431, mecA, mecR1, and mecI, which represent the genes coding for insertion sequence IS431, a modified penicillin binding protein 2a (PBP2a), the signal transducer protein, and the methicillin resistance regulatory protein (International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements, 2009; Shore and Coleman, 2013; Figure 1A).
Figure 1.
The psm-mec locus and its products. (A) The psm-mec gene is located on the class A mecA complex in SCCmec elements of types II, III, and VIII, next to the mecI/mecR1/mecA genes that confer methicillin resistance and its regulation. (B) The PSM-mec peptide product of the psm-mec locus forms an α-helix with pronounced amphiphathy; helical wheel presentation. (C) The psm-mec gene is part of a regulatory RNA with pronounced predicted secondary structure. The psm-mec gene is entirely embedded in a highly folded, large stem loop. The psm-mec srRNA only slightly extends toward the 3′ end and ends with a characteristic predicted terminator stem loop structure (Note, however, that Kaito et al. reported the srRNA to be slightly longer at the 5′ end). RBS, ribosomal binding site; Start codon coding for (N-formyl) methionine.
The psm-mec locus is found in the J2 region adjacent to mecI of the class A mec gene complex in several widespread HA-MRSA lineages, but most prominently in Sequence Type (ST) 5, ST36, ST45, ST225, and ST239 strains (Deurenberg and Stobberingh, 2009; Nübel et al., 2010; Castillo-Ramirez et al., 2012; Schulte et al., 2013), which all have either SCCmec types II or III (Chatterjee et al., 2011; Monecke et al., 2012; Josten et al., 2014). However, more detailed analysis revealed that the psm-mec locus is found in SCCmec types II, IIA, IIB, IID, III, and VIII of many Staphylococcus species (Monecke et al., 2012; Shore and Coleman, 2013).
Antibiotic resistance in bacteria is often associated with a reduced fitness cost to the bacterial host (Otto, 2013a). We recently reported that the expression of the PSM-mec peptide impacted oxacillin resistance in S. aureus (Cheung et al., 2014b), suggesting that a delicate balance may exist between virulence gene expression and methicillin resistance in MRSA strains carrying the psm-mec locus.
Expression and regulation of the PSM-mec peptide
The production of the PSM-mec peptide, which can be identified in bacterial culture filtrates by high-pressure liquid chromatography/mass spectrometry (HPLC/MS) (Queck et al., 2009) or Matrix-assisted laser desorption/ionization Time of Flight (MALDI-TOF) analysis (Josten et al., 2014), is a reliable indicator of methicillin resistance. However, the restricted distribution of the psm-mec locus severely limits the potential use of HPLC-based detection of PSM-mec for routine clinical screening of methicillin resistance.
The expression of all PSMs including the MGE-encoded PSM-mec is positively regulated by the accessory gene regulatory (agr) quorum sensing system (Novick, 2003; Queck et al., 2008; Chatterjee et al., 2011). This up-regulation is dependent on the binding of activated AgrA, the response regulator of agr, to promoter sequences upstream of the psm genes (Queck et al., 2008), but this has not directly been demonstrated for the psm-mec promoter. Interestingly, PSM expression is also enhanced in the presence of calf serum (Oogai et al., 2011) and up-regulated within intracellular environments after the activation of the stringent response (Geiger et al., 2012). Whether this is also the case for PSM-mec is not known.
PSM-mec production was shown to be especially frequent among clinical isolates of S. epidermidis compared to isolates from the skin of healthy individuals (Queck et al., 2009), which is a direct consequence of the genetic connection of psm-mec with methicillin resistance, which occurs much more frequently in clinical isolates. However, expression levels of PSM-mec can vary strongly among isolates (Queck et al., 2009). The mechanistic reasons for varying levels of PSM-mec expression between isolates cannot be fully explained but a documented -7T > C mutation in the psm-mec promoter is known to diminish PSM-mec expression in a subset of HA-MRSA strains (Kaito et al., 2011, 2013; Aoyagi et al., 2014).
Characterization of PSM-mec peptide
Like all members of the PSM family, PSM-mec is expressed with an N-terminal formyl-methionine and secreted without a signal peptide (Wang et al., 2007; Queck et al., 2009). Although this has not yet been shown directly for PSM-mec, it is likely that it is also exported by one or both of the two dedicated ATP-Binding Cassette (ABC) transporters that secrete PSMs (Chatterjee et al., 2013; Yoshikai et al., 2016). The 22 amino acid PSM-mec fits into the class of shorter α-type PSMs, which are anywhere between~ 20–25 amino acids in length, as opposed to the β-type, which are approximately twice as long (43–45 amino acids) (Cheung et al., 2014a).
PSM-mec has a unique cysteine residue at position 17, while cysteine residues are not present in the sequences of any other known PSM (Figure 1B). The notion of a hypothetical dimeric form of PSM-mec, due to the formation of disufide bridges created from oxidized cysteine residues, was formally discounted after size exclusion chromatography experiments showed no difference in extracted ion chromatograms of PSM-mec mutants compared to that of the unaltered PSM-mec peptide (Queck et al., 2009). However, the cysteine residue was crucial for pro-inflammatory and cytolytic activity, probably because it is essential for the PSM-mec secondary structure (Queck et al., 2009).
The pro-inflammatory and cytolytic properties of PSM-mec
At nanomolar concentrations, the PSMs are powerful pro-inflammatory agents and have an extraordinary capacity for activating and stimulating neutrophils (Wang et al., 2007), which is exerted through their interactions with human formyl peptide receptor 2 (FPR2; Kretschmer et al., 2010), a G protein-coupled receptor expressed on multiple immune cell types (Migeotte et al., 2006). Although differences in activity are observed between PSM members, PSM-mec readily up-regulates both CD11b and gp91phox expression, induces calcium flux, chemotaxis, and IL-8 release in neutrophils (Queck et al., 2009).
On the other hand, several PSM members of the α-type possess potent cytolytic activities, and an ever-increasing number of immune and non-immune cells are being discovered that are subject to PSM-mediated cytolysis (Cheung et al., 2014a). The lysis of eukaryotic cells by PSMs, which occurs at micromolar concentrations, is likely mediated in a receptor-independent manner (Kretschmer et al., 2010), While the involvement of receptors cannot be formally excluded, the facts that PSMs are surfactant molecules, lyse artificial phospholipid vesicles (Duong et al., 2012; Laabei et al., 2014), and FPR2 is not involved in lysis (Kretschmer et al., 2010), speak for such a receptor-independent mechanism. Lytic activity is, however, heavily influenced by membrane composition (Laabei et al., 2014). It is believed to be crucial for PSM-mediated immune evasion and progression of staphylococcal pathogenesis. PSM-mec follows the characteristic cytolytic profile of some α-type PSMs and can lyse human erythrocytes and neutrophils at a comparatively moderate level that is similar to that exerted by δ-toxin (Queck et al., 2009).
The psm-mec srRNA
The first clue for the presence of an srRNA within the psm-mec locus stemmed from experiments with MRSA strains transformed with a plasmid carrying 575 nucleotides amplified from SCCmec type II, termed the “F region” (Kaito et al., 2008). The authors reported that an open reading frame (ORF) transcribed in the opposite direction of the psm-mec gene, coined “fudoh,” encoded a protein that controlled colony spreading (Kaito et al., 2008). It was later discovered from experiments using S. aureus isogenic fudoh point-deletion mutants that the phenotypic differences were in fact influenced by an srRNA (Kaito et al., 2011), which contains the psm-mec gene (Kaito et al., 2011, 2013), in a way similar to the genetic layout of δ-toxin, whose gene is found within the regulatory RNA of the agr system, RNAIII (Novick, 2003). Interestingly, our group reported that the psm-mec srRNA transcript length was 143 bp, 14 nucleotides shorter than that described by Kaito et al. (2013) (Figure 1C). Notably, in any case, the psm-mec srRNA only barely exceeds the ORF coding for PSM-mec. This contrasts with RNAIII, which is considerably longer than the δ-toxin gene, hld. Additionally, the psm-mec srRNA is estimated to have a half-life of approximately 20 min (Kaito et al., 2013), which is significantly shorter than the ~45 min half-life described for RNAIII (Huntzinger et al., 2005). The only reported mechanism by which the psm-mec srRNA exerts it gene regulatory activity is through its interaction with the agrA transcript resulting in the overall decrease of AgrA activity (Kaito et al., 2013), which is entirely dependent on the first 60 nucleotides of the transcript (Kaito et al., 2013), where key anti-sense base-pair interactions occur (Cheung et al., 2014b). Because AgrA strictly regulates activity of the psmα and psmβ promoters (Queck et al., 2008), many of the psm-mec srRNA-mediated phenotypes likely are mostly due to a repression of the production of the strongly cytolytic and pro-inflammatory PSMα peptides (Table 1). Kaito et al. initially observed up to eight-fold reduction in PSM expression when the psm-mec locus was over-expressed on a plasmid in methicillin-sensitive S. aureus strains or strains with SCCmec type IV (Kaito et al., 2011). However, the differences in PSM reduction were less dramatic, and often inconsistent, in isogenic psm-mec mutants naturally harboring SCCmec types II and III (Chatterjee et al., 2011), indicating a high degree of strain dependence regarding the role of the psm-mec srRNA in S. aureus.
Table 1.
Activities of the PSM-mec peptide and the psm-mec srRNA.
| Virulence determinant within the psm-mec locus | Known in vitro activities in S. aureus |
|---|---|
| PSM-mec peptide | FPR2-mediated pro-inflammatory activity at nM concentrations: • Increased CD11b expression • Increased gp91phox expression • Increased IL-8 expression • Increased calcium flux • Increased chemotaxis |
| Receptor-independent cytolytic activity at μM concentrations toward: • Erythrocytes • Neutrophils |
|
| Peptide expression decreases oxacillin resistancea | |
| psm-mec srRNA | Represses AgrA activity through direct interaction • Decreased PSM productionb • Enhanced biofilm formation and aggregationb • Suppresses colony spreading • Up-regulates SpA expressionb |
Only tested in one background strain.
Strain-specific phenotype.
The impact of the psm-mec srRNA on biofilm formation, colony spreading, and expression of S. aureus virulence factors
PSM peptides have a considerable impact on biofilm formation, which is based on their detergent-like properties (Vuong et al., 2000; Kong et al., 2006). PSMs cause the structuring and maturation of biofilms, which includes the formation of nutrient-transporting channels (Wang et al., 2011; Periasamy et al., 2012). PSMs also facilitate the detachment of biofilm clusters in vitro and in vivo (Wang et al., 2011; Periasamy et al., 2012; Otto, 2013b). It was shown that isogenic agr mutants (Vuong et al., 2000, 2004), total psm deletion mutants in all psm genes, and even single mutants in either the psmα, psmβ, or hld loci (Wang et al., 2011; Periasamy et al., 2012) formed thicker biofilms compared to their wild type counterparts. Therefore, it was surprising to find that several isogenic psm-mec S. aureus mutants showed a reverse phenotype, albeit the change in biofilm formation was overall minor (Queck et al., 2009; Kaito et al., 2013). A phenotype of increased aggregation similar to that conferred by other PSMs (Dastgheyb et al., 2015) was only found in a strain in which the PSM-mec peptide was expressed in high relative amounts as compared to other PSMs (Queck et al., 2009). In most psm-mec-positive S. aureus strains, the regulatory effect of the psm-mec srRNA on the expression of other biofilm-dispersing PSMs (Periasamy et al., 2012) appears to over-shadow the lack of the direct dispersion effect of the PSM-mec peptide in psm-mec mutants (Kaito et al., 2013). Thus, in most strains, the impact of the psm-mec locus on biofilm formation is opposite to that of other psm loci, although that effect is generally not very pronounced.
For many pathogenic bacteria, motility plays an important role in colonization and virulence (Cossart, 2002; Josenhans and Suerbaum, 2002; Krukonis and DiRita, 2003). Motility is commonly dependent on the expression of flagella and type IV pili (Kearns, 2010). Even though S. aureus has always been historically regarded as non-motile, as it does not have the genes for flagella or pili, S. aureus has repeatedly been shown to spread efficiently across the surface of soft agar (Kaito et al., 2011; Tsompanidou et al., 2012) and more recently, on fresh pork meat (Tsompanidou et al., 2013), through a PSM-dependent mechanism (Lin et al., 2016), an effect now known as “colony-spreading” (Kaito and Sekimizu, 2007).
Originally, the observations of colony spreading and several other phenotypes were made using plasmid-based expression of the psm-mec locus in MRSA strains that did not carry the psm-mec locus, such as S. aureus isolates that were either methicillin-sensitive or MRSA strains that have recently emerged in the community (Community-Associated MRSA, CA-MRSA (Lowy, 1998; DeLeo et al., 2010). CA-MRSA strains are genetically distinct and notably more aggressive than HA-MRSA strains, show enhanced production of virulence factors, and are capable of causing disease in otherwise healthy individuals outside public healthcare settings (Tacconelli et al., 2004; Diep and Otto, 2008; Li et al., 2009; DeLeo et al., 2010). Furthermore, CA-MRSA strains characteristically possess SCCmec types IV or V (Diep and Otto, 2008; DeLeo et al., 2010) and therefore lack the psm-mec locus. Interestingly, CA-MRSA strains were reported to have a greater capacity to spread on soft agar compared to HA-MRSA strains (Kaito et al., 2008) and the absence of the psm-mec locus from CA-MRSA strains was speculated to cause that phenotype. Indeed, CA-MRSA strains transformed with plasmids containing the psm-mec locus were impaired in colony spreading. Conversely, isogenic HA-MRSA psm-mec mutants showed enhanced colony spreading (Kaito et al., 2013). Moreover, the inability of synthetic PSM-mec to rescue the colony spreading phenotype in an agr deletion strain of S. aureus (Tsompanidou et al., 2013) indicated that this regulation is predominantly mediated by the psm-mec srRNA. However, other studies showed that colony spreading is positively regulated by agr (Tsompanidou et al., 2011) and mainly controlled by mechanisms independent of the psm-mec srRNA, which include the production of several staphylococcal virulence factors, such as PSMs (Omae et al., 2012; Tsompanidou et al., 2013) and teichoic acids (Kaito and Sekimizu, 2007). In contrast, colony spreading is suppressed in the presence of multiple staphylococcal cell surface proteins (Tsompanidou et al., 2012). Taken together, the molecular mechanisms behind colony spreading in S. aureus reveal a somewhat complex network involving different, and often opposing, signals that affect the final phenotypic outcome. Lastly, it remains unknown whether colony spreading on agar surface is a mechanism universally adopted by all staphylococci and to what extent it matters for the colonization of epithelial surfaces in vivo.
In an effort to investigate psm-mec-dependent regulation in the natural strain background without artificial plasmid expression-induced effects, our group used microarray profiling to compare the transcriptomes of wild-type strains compared with those of isogenic psm-mec mutants (Cheung et al., 2014b). Most strikingly, the primary conserved target of psm-mec-dependent gene regulation was spa, the gene coding for the immune evasion protein, surface protein A (SpA), which was independently confirmed (Kaito et al., 2011). We also found that the strength of psm-mec srRNA transcription affected SpA expression (Cheung et al., 2014b). The up-regulation of SpA by the psm-mec srRNA is especially interesting because it adds to a number of existing transcriptional regulators known to control spa (Gao and Stewart, 2004). The up-regulated expression of SpA may play an important but yet unidentified role in the manifestation of staphylococcal disease (Gomez et al., 2004; Cheng et al., 2009; Merino et al., 2009; Kim et al., 2011) in some psm-mec-harboring S. aureus strains.
The impact of the psm-mec locus on S. aureus virulence
Our group demonstrated that the ability of an isogenic psm-mec S. aureus mutant to form skin abscesses and cause sepsis in mouse models of infection was severely attenuated compared to the parent strain (Queck et al., 2009), indicating that the PSM-mec peptide contributed significantly to S. aureus pathogenesis. However, this phenotype could only be produced using an isogenic psm-mec S. aureus mutant whose parent strain produced more PSM-mec relative to other PSMs as compared to most psm-mec-harboring strains. Kaito et al. reported that the psm-mec srRNA contributes to virulence, but in a fashion opposite to that mediated by the PSM-mec peptide (Kaito et al., 2011, 2013). Furthermore, those authors proposed that the psm-mec srRNA was generally responsible for the differences in virulence between HA- and CA-MRSA strains (Kaito et al., 2013). They also speculated that point mutations in the psm-mec locus underlie increased virulence of a certain lineage, ST764, but this was not directly shown using isogenic mutants (Suzuki et al., 2016). However, a phenotype of increased virulence is not always observed in isogenic psm-mec HA-MRSA strains (Chatterjee et al., 2011), suggesting that the contribution of psm-mec srRNA toward S. aureus pathogenesis is strongly strain-dependent and cannot serve as a generally applicable explanation for differences in virulence between CA- and HA-MRSA strains.
The roles of the PSM-mec peptide and the psm-mec regulatory RNA in other Staphylococcus species
In addition to S. aureus, the psm-mec locus is detected in many different methicillin-resistant Staphylococcus species (Monecke et al., 2012). However, PSM-mec expression levels have only been rigorously studied in S. epidermidis (Queck et al., 2009). Our group showed that PSM-mec expression was only detected in methicillin-resistant S. epidermidis infection isolates, but never in methicillin-sensitive S. epidermidis strains (Queck et al., 2009). There is no amino acid or nucleotide sequence difference (100% identity) between the S. aureus and S. epidermidis PSM-mec peptide or srRNA, respectively, based on a comparison of strains S. aureus MRSA252 and S. epidermidis RP62A, and differences are also absent or very minor in other strains. This is in accordance with the notion of a relatively recent move of SCCmec from coagulase-negative species to S. aureus.
To date, only one study has studied the impact of the psm-mec locus on the phenotypes in S. epidermidis and S. haemolyticus (Ikuo et al., 2014). The results, which were based on S. epidermidis and S. haemolyticus strains without SCCmec transformed with plasmids harboring the psm-mec locus, showed a decrease in PSM production, as demonstrated in similar experiments with S. aureus (Kaito et al., 2011). Intriguingly, an increase of biofilm formation was only observed in experiments with S. epidermidis, as reported previously (Kaito et al., 2011), but not S. haemolyticus, suggesting that the impact of the psm-mec locus on virulence phenotypes among staphylococci is also highly strain-dependent. The role of PSM-mec in coagulase-negative staphylococci has not yet been directly investigated using isogenic deletion mutants.
Conclusion
It has become clear that the effects exerted by the psm-mec locus in S. aureus on virulence and virulence phenotypes such as biofilm formation are highly strain-dependent. They are also commonly rather moderate in extent compared to those conferred by other members of the PSM family. While great strides have been made to better understand the molecular underpinnings of gene regulatory psm-mec-dependent mechanisms in S. aureus pathogenesis, in the future the focus should be shifted toward the investigation of how the psm-mec locus affects the pathogenesis in other Staphylococcus species, especially S. epidermidis and S. haemolyticus, which, alongside S. aureus, are the most frequent and pathogenic agents of hospital-associated infections around the world. As it is a commonly accepted notion that SCCmec elements originate from coagulase-negative staphylococci (Otto, 2013c), such investigation is bound to shed light on the “original” role of the psm-mec locus in staphylococcal physiology, with the potential to also explain the contrasting roles of psm-mec in the S. aureus background.
Author contributions
LQ, JM, GC, and MO contributed to the drafting of the manuscript and approved the final version.
Funding
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, U.S. National Institutes of Health (grant ZIA AI000904-16).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Aoyagi T., Kaito C., Sekimizu K., Omae Y., Saito Y., Mao H., et al. (2014). Impact of psm-mec in the mobile genetic element on the clinical characteristics and outcome of SCCmec-II methicillin-resistant Staphylococcus aureus bacteraemia in Japan. Clin. Microbiol. Infect. 20, 912–919. 10.1111/1469-0691.12575 [DOI] [PubMed] [Google Scholar]
- Cassat J. E., Hammer N. D., Campbell J. P., Benson M. A., Perrien D. S., Mrak L. N., et al. (2013). A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe 13, 759–772. 10.1016/j.chom.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Ramirez S., Corander J., Marttinen P., Aldeljawi M., Hanage W. P., Westh H., et al. (2012). Phylogeographic variation in recombination rates within a global clone of methicillin-resistant Staphylococcus aureus. Genome Biol. 13:R126. 10.1186/gb-2012-13-12-r126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S. S., Chen L., Joo H. S., Cheung G. Y., Kreiswirth B. N., Otto M. (2011). Distribution and regulation of the mobile genetic element-encoded phenol-soluble modulin PSM-mec in methicillin-resistant Staphylococcus aureus. PLoS ONE 6:e28781. 10.1371/journal.pone.0028781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S. S., Joo H. S., Duong A. C., Dieringer T. D., Tan V. Y., Song Y., et al. (2013). Essential Staphylococcus aureus toxin export system. Nat. Med. 19, 364–367. 10.1038/nm.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. G., Kim H. K., Burts M. L., Krausz T., Schneewind O., Missiakas D. M. (2009). Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 23, 3393–3404. 10.1096/fj.09-135467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung G. Y., Joo H. S., Chatterjee S. S., Otto M. (2014a). Phenol-soluble modulins–critical determinants of staphylococcal virulence. FEMS Microbiol. Rev. 38, 698–719. 10.1111/1574-6976.12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung G. Y., Villaruz A. E., Joo H. S., Duong A. C., Yeh A. J., Nguyen T. H., et al. (2014b). Genome-wide analysis of the regulatory function mediated by the small regulatory psm-mec RNA of methicillin-resistant Staphylococcus aureus. Int. J. Med. Microbiol. 304, 637–644. 10.1016/j.ijmm.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P. (2002). Molecular and cellular basis of the infection by Listeria monocytogenes: an overview. Int. J. Med. Microbiol. 291, 401–409. 10.1078/1438-4221-00146 [DOI] [PubMed] [Google Scholar]
- Costerton J. W. (1999). Introduction to biofilm. Int. J. Antimicrob. Agents 11, 217–221. discussion: 237–239. [DOI] [PubMed] [Google Scholar]
- Dastgheyb S. S., Villaruz A. E., Le K. Y., Tan V. Y., Duong A. C., Chatterjee S. S., et al. (2015). Role of phenol-soluble modulins in formation of Staphylococcus aureus biofilms in synovial fluid. Infect. Immun. 83, 2966–2975. 10.1128/IAI.00394-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo F. R., Otto M., Kreiswirth B. N., Chambers H. F. (2010). Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375, 1557–1568. 10.1016/S0140-6736(09)61999-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurenberg R. H., Stobberingh E. E. (2009). The molecular evolution of hospital- and community-associated methicillin-resistant Staphylococcus aureus. Curr. Mol. Med. 9, 100–115. 10.1111/j.1469-0691.2006.01573.x [DOI] [PubMed] [Google Scholar]
- Diep B. A., Otto M. (2008). The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 16, 361–369. 10.1016/j.tim.2008.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong A. C., Cheung G. Y., Otto M. (2012). Interaction of phenol-soluble modulins with phosphatidylcholine vesicles. Pathogens 1, 3–11. 10.3390/pathogens1010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Stewart G. C. (2004). Regulatory elements of the Staphylococcus aureus protein A (Spa) promoter. J. Bacteriol. 186, 3738–3748. 10.1128/JB.186.12.3738-3748.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T., Francois P., Liebeke M., Fraunholz M., Goerke C., Krismer B., et al. (2012). The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 8:e1003016. 10.1371/journal.ppat.1003016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez M. I., Lee A., Reddy B., Muir A., Soong G., Pitt A., et al. (2004). Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat. Med. 10, 842–848. 10.1038/nm1079 [DOI] [PubMed] [Google Scholar]
- Huntzinger E., Boisset S., Saveanu C., Benito Y., Geissmann T., Namane A., et al. (2005). Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24, 824–835. 10.1038/sj.emboj.7600572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuo M., Nagano G., Saito Y., Mao H., Sekimizu K., Kaito C. (2014). Inhibition of exotoxin production by mobile genetic element SCCmec-encoded psm-mec RNA is conserved in staphylococcal species. PLoS ONE 9:e100260. 10.1371/journal.pone.0100260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Okuma K., Ma X. X., Yuzawa H., Hiramatsu K. (2003). Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Updat. 6, 41–52. 10.1016/S1368-7646(03)00003-7 [DOI] [PubMed] [Google Scholar]
- International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements . (2009). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53, 4961–4967. 10.1128/AAC.00579-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josenhans C., Suerbaum S. (2002). The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291, 605–614. 10.1078/1438-4221-00173 [DOI] [PubMed] [Google Scholar]
- Josten M., Dischinger J., Szekat C., Reif M., Al-Sabti N., Sahl H., et al. (2014). Identification of agr-positive methicillin-resistant Staphylococcus aureus harbouring the class A mec complex by MALDI-TOF mass spectrometry. Int. J. Med. Microbiol. 304, 1018–1023. 10.1016/j.ijmm.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Kaito C., Omae Y., Matsumoto Y., Nagata M., Yamaguchi H., Aoto T., et al. (2008). A novel gene, fudoh, in the SCCmec region suppresses the colony spreading ability and virulence of Staphylococcus aureus. PLoS ONE 3:e3921. 10.1371/journal.pone.0003921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaito C., Saito Y., Ikuo M., Omae Y., Mao H., Nagano G., et al. (2013). Mobile genetic element SCCmec-encoded psm-mec RNA suppresses translation of agrA and attenuates MRSA virulence. PLoS Pathog. 9:e1003269. 10.1371/journal.ppat.1003269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaito C., Saito Y., Nagano G., Ikuo M., Omae Y., Hanada Y., et al. (2011). Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence. PLoS Pathog. 7:e1001267. 10.1371/journal.ppat.1001267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaito C., Sekimizu K. (2007). Colony spreading in Staphylococcus aureus. J. Bacteriol. 189, 2553–2557. 10.1128/JB.01635-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8, 634–644. 10.1038/nrmicro2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K., Kim H. Y., Schneewind O., Missiakas D. (2011). Identifying protective antigens of Staphylococcus aureus, a pathogen that suppresses host immune responses. FASEB J. 25, 3605–3612. 10.1096/fj.11-187963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong K. F., Vuong C., Otto M. (2006). Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 296, 133–139. 10.1016/j.ijmm.2006.01.042 [DOI] [PubMed] [Google Scholar]
- Kretschmer D., Gleske A. K., Rautenberg M., Wang R., Köberle M., Bohn E., et al. (2010). Human formyl peptide receptor 2 senses highly pathogenic Staphylococcus aureus. Cell Host Microbe 7, 463–473. 10.1016/j.chom.2010.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukonis E. S., DiRita V. J. (2003). From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6, 186–190. 10.1016/S1369-5274(03)00032-8 [DOI] [PubMed] [Google Scholar]
- Laabei M., Jamieson W. D., Yang Y., van den Elsen J., Jenkins A. T. (2014). Investigating the lytic activity and structural properties of Staphylococcus aureus phenol soluble modulin (PSM) peptide toxins. Biochim. Biophys. Acta 1838, 3153–3161. 10.1016/j.bbamem.2014.08.026 [DOI] [PubMed] [Google Scholar]
- Li M., Diep B. A., Villaruz A. E, Braughton K. R, Jiang X., DeLeo F. R., et al. (2009). Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 5883–5888. 10.1073/pnas.0900743106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Skov R. L., Han X., Larsen A. R., Larsen J., Sørum M., et al. (2011). Novel types of staphylococcal cassette chromosome mec elements identified in clonal complex 398 methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 55, 3046–3050. 10.1128/AAC.01475-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. H., Ke W. J., Liu C. C., Yang M. W. (2016). Modulation of Staphylococcus aureus spreading by water. Sci. Rep. 6:25233. 10.1038/srep25233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- Merino N., Toledo-Arana A., Vergara-Irigaray M., Valle J., Solano C., Calvo E., et al. (2009). Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 191, 832–843. 10.1128/JB.01222-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeotte I., Communi D., Parmentier M. (2006). Formyl peptide receptors: a promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 17, 501–519. 10.1016/j.cytogfr.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Monecke S., Engelmann I., Archambault M., Coleman D. C., Coombs G. W., Cortez de Jäckel S., et al. (2012). Distribution of SCCmec-associated phenol-soluble modulin in staphylococci. Mol. Cell. Probes 26, 99–103. 10.1016/j.mcp.2012.01.001 [DOI] [PubMed] [Google Scholar]
- Novick R. P. (2003). Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48, 1429–1449. 10.1046/j.1365-2958.2003.03526.x [DOI] [PubMed] [Google Scholar]
- Nübel U., Dordel J., Kurt K., Strommenger B., Westh H., Shukla S., et al. (2010). A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-resistant Staphylococcus aureus. PLoS Pathog. 6:e1000855. 10.1371/journal.ppat.1000855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omae Y., Sekimizu K., Kaito C. (2012). Inhibition of colony-spreading activity of Staphylococcus aureus by secretion of delta-hemolysin. J. Biol. Chem. 287, 15570–15579. 10.1074/jbc.M112.357848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oogai Y., Matsuo M., Hashimoto M., Kato F., Sugai M., Komatsuzawa H. (2011). Expression of virulence factors by Staphylococcus aureus grown in serum. Appl. Environ. Microbiol. 77, 8097–8105. 10.1128/AEM.05316-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2008). Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322, 207–228. 10.1007/978-3-540-75418-3_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2009). Staphylococcus epidermidis–the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7, 555–567. 10.1038/nrmicro2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2012). MRSA virulence and spread. Cell. Microbiol. 14, 1513–1521. 10.1111/j.1462-5822.2012.01832.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2013a). Community-associated MRSA: what makes them special? Int. J. Med. Microbiol. 303, 324–330. 10.1016/j.ijmm.2013.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2013b). Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu. Rev. Med. 64, 175–188. 10.1146/annurev-med-042711-140023 [DOI] [PubMed] [Google Scholar]
- Otto M. (2013c). Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays 35, 4–11. 10.1002/bies.201200112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2014). Staphylococcus aureus toxins. Curr. Opin. Microbiol. 17, 32–37. 10.1016/j.mib.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy S., Joo H. S., Duong A. C., Bach T. H., Tan V. Y., Chatterjee S. S., et al. (2012). How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1281–1286. 10.1073/pnas.1115006109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queck S. Y., Jameson-Lee M., Villaruz A. E., Bach T. H., Khan B. A., Sturdevant D. E., et al. (2008). RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32, 150–158. 10.1016/j.molcel.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queck S. Y., Khan B. A., Wang R., Bach T. H., Kretschmer D., Chen L., et al. (2009). Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog. 5:e1000533. 10.1371/journal.ppat.1000533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte B., Bierbaum G., Pohl K., Goerke C., Wolz C. (2013). Diversification of clonal complex 5 methicillin-resistant Staphylococcus aureus strains (Rhine-Hesse clone) within Germany. J. Clin. Microbiol. 51, 212–216. 10.1128/JCM.01967-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore A. C., Coleman D. C. (2013). Staphylococcal cassette chromosome mec: recent advances and new insights. Int. J. Med. Microbiol. 303, 350–359. 10.1016/j.ijmm.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Shore A. C., Deasy E. C., Slickers P., Brennan G., O'Connell B., Monecke S., et al. (2011). Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 55, 3765–3773. 10.1128/AAC.00187-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Yamamoto T., Kaito C., Miyamoto H., Ohashi Y. (2016). Impact of psm-mec in methicillin-resistant Staphylococcus aureus (ST764) strains isolated from keratitis patients. Microb. Drug Resist. 22, 137–140. 10.1089/mdr.2015.0315 [DOI] [PubMed] [Google Scholar]
- Tacconelli E., Venkataraman L., De Girolami P. C, DAgata E. M. (2004). Methicillin-resistant Staphylococcus aureus bacteraemia diagnosed at hospital admission: distinguishing between community-acquired versus healthcare-associated strains. J. Antimicrob. Chemother. 53, 474–479. 10.1093/jac/dkh107 [DOI] [PubMed] [Google Scholar]
- Tsompanidou E., Denham E. L., Becher D., de Jong A., Buist G., van Oosten M., et al. (2013). Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfaces. Appl. Environ. Microbiol. 79, 886–895. 10.1128/AEM.03157-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsompanidou E., Denham E. L., Sibbald M. J., Yang X. M., Seinen J., Friedrich A. W., et al. (2012). The sortase A substrates FnbpA, FnbpB, ClfA and ClfB antagonize colony spreading of Staphylococcus aureus. PLoS ONE 7:e44646. 10.1371/journal.pone.0044646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsompanidou E., Sibbald M. J., Chlebowicz M. A., Dreisbach A., Back J. W., van Dijl J. M., et al. (2011). Requirement of the agr locus for colony spreading of Staphylococcus aureus. J. Bacteriol. 193, 1267–1272. 10.1128/JB.01276-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C., Kocianova S., Yao Y., Carmody A. B., Otto M. (2004). Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190, 1498–1505. 10.1086/424487 [DOI] [PubMed] [Google Scholar]
- Vuong C., Saenz H. L., Götz F., Otto M. (2000). Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182, 1688–1693. 10.1086/317606 [DOI] [PubMed] [Google Scholar]
- Wang R., Braughton K. R., Kretschmer D., Bach T. H., Queck S. Y., Li M., et al. (2007). Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13, 1510–1514. 10.1038/nm1656 [DOI] [PubMed] [Google Scholar]
- Wang R., Khan B. A., Cheung G. Y., Bach T. H., Jameson-Lee M., Kong K. F., et al. (2011). Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Invest. 121, 238–248. 10.1172/JCI42520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikai H., Kizaki H., Saito Y., Omae Y., Sekimizu K., Kaito C. (2016). Multidrug-resistance transporter AbcA secretes Staphylococcus aureus cytolytic toxins. J. Infect. Dis. 213, 295–304. 10.1093/infdis/jiv376 [DOI] [PubMed] [Google Scholar]