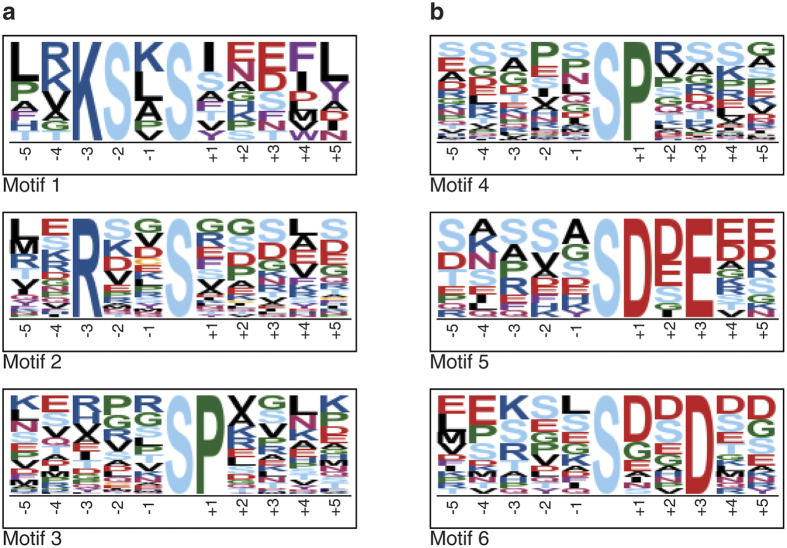
Figure 3. Identification of potential in vivo SnRK1 substrate candidates.
Motif-x50 analysis was done for phosphopeptides that were at least 50% more abundant or significantly upregulated in the wild type (a) and in snrk1α1/α2 mutant (b). p-value threshold was set to 0.000001 and at least 10 occurences were required. IPI Arabidopsis proteome was used as background proteome. For further details see Results and Discussion.