A copper(II) complex of a pyridine-containing macrocycle (PyMAC) reveals a six-coordinated octahedral CuII complex with a tetradentate aminopyridine macrocycle ligand surrounding the metal centre in a square-planar geometry. Two weakly bound perchlorate counter-ions occupy the axial sites above and below the macrocyclic plane.
Keywords: crystal structure, pyridine macrocycles, copper complex
Abstract
The title copper(II) complex of a pyridine-containing macrocycle (PyMAC), [Cu(C16H28N4)](ClO4)2, has been prepared. The crystal structure reveals the CuII atom to be octahedrally coordinated by a tetradentate aminopyridine macrocyclic ligand surrounding the metal cation in a square-planar geometry. Two weakly bound perchlorate counter-ions occupy the axial sites above and below the macrocyclic plane. The crystal studied was refined as a two-component pseudo-merohedral twin; the refined fractional contribution of the minor component is 38.77 (8)%
Chemical context
There have been several studies of the macrocycles synthesized from 2,6-diacetylpyridine and polyamines. One of the first examples, reported by Karn & Busch (1966 ▸), involved a nickel(II)-templated condensation of 2,6-diacetylpyridine and bis(3-aminopropyl)amine. Their pioneering work enabled subsequent syntheses of various pyridine-containing macrocycles (Rezaeivala & Keypour, 2014 ▸), including a family of complexes with appended arms (PyMACs) (Organo et al., 2009 ▸; Herrera et al., 2003 ▸) as shown in Fig. 1 ▸.
Figure 1.
Pyridine-containing macrocycles (PyMACs).
Various metal ions have been incorporated into PyMAC ligands, and the resulting complexes often showed interesting catalytic properties. For example, NiII–PyMAC complexes have been found to exhibit peroxidase-like activity, with NiLCOOH (Fig. 1 ▸) being most active (Organo et al., 2009 ▸). FeII–LMe (Fig. 1 ▸) was also found to have catalytic use in epoxidation reactions of cyclooctene with hydrogen peroxide (Ye et al., 2012 ▸). A similar CuII–PyMAC complex but without methyl groups at the macrocyclic ring was reported by Fernandes et al. (2007 ▸) to scavenge superoxide.
Pyridine-containing metallomacrocycles have also found utility beyond synthetic chemistry. For example, Cu–macrocyclic complexes have become increasingly important in radiopharmaceutical applications as contrast agents in positron emission tomographic (PET) imaging (Boros et al., 2014 ▸).
While there are known Cu–pyridine macrocycles, only a few have been characterized structurally (Caira et al., 1975 ▸; Lindoy et al., 2001 ▸; Herrera et al., 2003 ▸; Autzen et al., 2003 ▸). Here, we report the synthesis and crystal structure of a CuII–PyMAC perchlorate compound.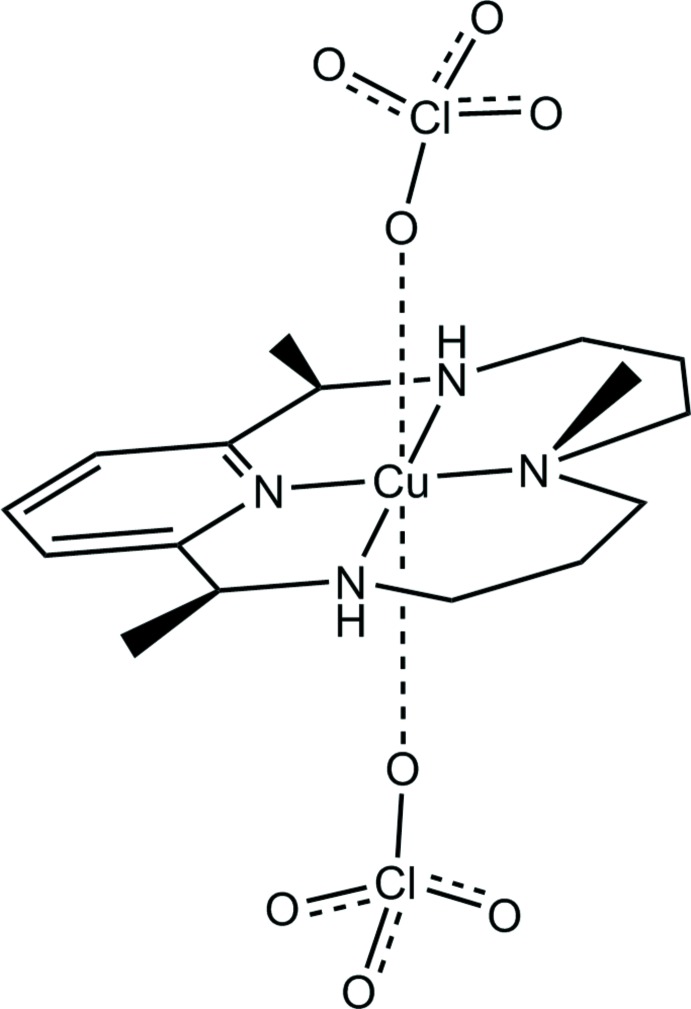
Structural commentary
The title compound has the CuII atom in a distorted octahedral coordination, with the tetradentate aminopyridine macrocyclic ligand surrounding the metal atom in a square-planar geometry (Fig. 2 ▸). Two perchlorate counter-ions occupy the axial sites perpendicular to the macrocyclic plane. The macrocyclic ligand incorporates a 2,6-substituted pyridine unit that is connected on both sides to an aliphatic chain of 11 atoms, including two secondary amines and a tertiary amine bearing a methyl group. When coordinated to the CuII atom, the macrocycle exhibits approximate molecular mirror symmetry with respect to the plane that bisects the pyridine and tertiary amine nitrogen atoms, and is perpendicular to the macrocyclic plane. The Cu—N distances between CuII and secondary amine nitrogen atoms [2.0417 (14) and 2.0445 (15) Å] are similar to each other; the distance between CuII and the tertiary amine N atom [2.0108 (13) Å] is slightly shorter. In contrast, the Cu—Npy bond length [1.9316 (13) Å] is much shorter than the Cu—Namine bonds. Both perchlorate anions are only weakly bound, with Cu—O6 and Cu—O3 distances of 2.6478 (13) and 2.4736 (13) Å, respectively.
Figure 2.
An ORTEP diagram of the molecular structure of CuLMe(ClO4)2 [LMe = 2,7,12-trimethyl-3,7,11,17-tetraazabicyclo[11.3.1]heptadeca-1(17),13,15-triene, see Fig. 1 ▸], showing the atom-labeling scheme, with ellipsoids drawn at the 50% probability level. Hydrogen atoms are omitted for clarity.
An intramolecular contact (N4—H4⋯O5) occurs between a perchlorate O atom and the tertiary amine NH group. The N⋯O distance [3.423 (2) Å] is longer than the sum of van der Waals radii of the two atoms (2.94 Å), suggesting this is a weaker interaction comparing to normal hydrogen-bonding interactions.
Supramolecular features
In the crystal of the complex (see Fig. 3 ▸), several N—H⋯O and Cpy—H⋯O hydrogen bonds have longer D⋯A distances than the van der Waals radii of the corresponding pairs of atoms (3.25 Å for C⋯O). The resulting geometry is a chain along [010]. Numerical details are given in Table 1 ▸.
Figure 3.
Crystal packing of the title complex viewed approximately down the a axis. Hydrogen atoms, except those involved in hydrogen bonds, are omitted for clarity.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2⋯O7i | 0.83 (2) | 2.94 (2) | 3.536 (2) | 130.6 (18) |
| N4—H4⋯O4ii | 0.86 (2) | 2.45 (2) | 3.1619 (19) | 140.2 (18) |
| N4—H4⋯O5 | 0.86 (2) | 2.77 (2) | 3.423 (2) | 134.1 (17) |
| C2—H2A⋯O5iii | 0.93 | 2.70 | 3.587 (2) | 161 |
| C3—H3⋯O1iv | 0.93 | 2.68 | 3.585 (2) | 165 |
| C4—H4A⋯O1v | 0.93 | 2.64 | 3.518 (2) | 158 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Synthesis and crystallization
The procedure for the synthesis of the title compound was adapted from Karn & Busch (1966 ▸) with subsequent reduction using NaBH4. 10 mmol of 2,6-diacetylpyridine were dissolved in 160 ml of absolute ethanol, and the resulting solution was mixed with 10 mmol of Cu(ClO4)2·6H2O in 240 ml of water. The reaction mixture was heated to 338 K and 10 mmol of N,N-bis(3-aminopropyl)methylamine were added. Subsequently, glacial acetic acid was added to the mixture until the pH was about 4. The mixture was heated to reflux of the solvent for 12 h; a color change from blue to dark blue occurred during that period. After reflux, the mixture was cooled to room temperature and 40 mmol of NaBH4 were added. The mixture was left to stir for 12 h for complete reduction. Perchloric acid was added until the remaining NaBH4 was consumed.
The deep-blue solution was concentrated to about a tenth of its original volume by rotary evaporation. The solution was then cooled slowly to room temperature and refrigerated. Dark-purple needle-like crystals formed upon cooling. The crystals were filtered, washed with absolute ethanol and diethyl ether, and allowed to dry. Light-purple crystals were recrystallized from hot water. Single crystals were obtained by dissolving the compound in acetonitrile followed by slow ether diffusion.
UV–Vis data: λmax = 552 nm in methanol, molar extinction coefficient: 209.47 M −1·cm−1. IR: 1619 cm−1 (C=N of pyridine), 1113 and 600 cm−1 (ClO4 − bands) and 3400 cm−1 (N—H).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The crystal structure was refined as a two-component pseudo-merohedral twin (twin operation:  00, 0
00, 0 0, 001); the refined fractional contribution of the minor component is 38.77 (8)%. All H atoms bonded to C atoms were placed at calculated positions using a riding model, with C—H distances of 0.98 Å for CH, 0.97 Å for CH2, 0.96 Å for CH3, and 0.93 Å for aromatic CH, and with U
iso(H) = 1.2U
eq(C) for all but CH3 where U
iso(H) = 1.5U
eq(C). H2 and H4 connected to N2 and N4 were located in the difference density Fourier synthesis maps and refined freely.
0, 001); the refined fractional contribution of the minor component is 38.77 (8)%. All H atoms bonded to C atoms were placed at calculated positions using a riding model, with C—H distances of 0.98 Å for CH, 0.97 Å for CH2, 0.96 Å for CH3, and 0.93 Å for aromatic CH, and with U
iso(H) = 1.2U
eq(C) for all but CH3 where U
iso(H) = 1.5U
eq(C). H2 and H4 connected to N2 and N4 were located in the difference density Fourier synthesis maps and refined freely.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | [Cu(C16H28N4)](ClO4)2 |
| M r | 538.86 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 100 |
| a, b, c (Å) | 8.6918 (12), 12.0588 (16), 20.068 (3) |
| β (°) | 90.153 (3) |
| V (Å3) | 2103.4 (5) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 1.35 |
| Crystal size (mm) | 0.24 × 0.21 × 0.21 |
| Data collection | |
| Diffractometer | Bruker D8 QUEST |
| Absorption correction | Multi-scan (Krause et al., 2015 ▸) |
| T min, T max | 0.414, 0.454 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 36478, 5284, 5054 |
| R int | 0.030 |
| (sin θ/λ)max (Å−1) | 0.687 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.026, 0.062, 1.06 |
| No. of reflections | 5284 |
| No. of parameters | 292 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.50, −0.33 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016009701/zl2666sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016009701/zl2666Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016009701/zl2666sup3.tif
CCDC reference: 1485717
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
Financial support by the National Science Foundation (CHE14129090 and CHE1229426) and the UP System Emerging Interdisciplinary Research Program (OVPAA-EIDR 12-001-121102) is greatly acknowledged.
supplementary crystallographic information
Crystal data
| [Cu(C16H28N4)](ClO4)2 | F(000) = 1116 |
| Mr = 538.86 | Dx = 1.702 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.6918 (12) Å | Cell parameters from 9732 reflections |
| b = 12.0588 (16) Å | θ = 2.9–30.6° |
| c = 20.068 (3) Å | µ = 1.35 mm−1 |
| β = 90.153 (3)° | T = 100 K |
| V = 2103.4 (5) Å3 | Clear dark blue cube, clear dark blue |
| Z = 4 | 0.24 × 0.21 × 0.21 mm |
Data collection
| Bruker D8 QUEST diffractometer | 5284 independent reflections |
| Radiation source: sealed tube | 5054 reflections with I > 2σ(I) |
| Detector resolution: 1.024 pixels mm-1 | Rint = 0.030 |
| φ and ω scans | θmax = 29.2°, θmin = 2.9° |
| Absorption correction: multi-scan (Krause et al., 2015) | h = −11→11 |
| Tmin = 0.414, Tmax = 0.454 | k = −16→16 |
| 36478 measured reflections | l = −27→27 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.026 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.062 | w = 1/[σ2(Fo2) + (0.027P)2 + 0.369P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max = 0.002 |
| 5284 reflections | Δρmax = 0.50 e Å−3 |
| 292 parameters | Δρmin = −0.33 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refined as a 2-component twin. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cu1 | 0.19755 (3) | 0.24103 (2) | 0.38162 (2) | 0.01124 (5) | |
| N1 | 0.00263 (15) | 0.31793 (10) | 0.38014 (7) | 0.0121 (2) | |
| N2 | 0.14981 (17) | 0.22614 (12) | 0.48078 (7) | 0.0131 (3) | |
| H2 | 0.131 (3) | 0.1590 (18) | 0.4820 (11) | 0.016 (5)* | |
| N3 | 0.40687 (15) | 0.17015 (10) | 0.38456 (7) | 0.0133 (2) | |
| N4 | 0.16917 (17) | 0.24951 (11) | 0.28059 (7) | 0.0133 (3) | |
| H4 | 0.126 (2) | 0.1870 (18) | 0.2721 (10) | 0.014 (5)* | |
| C1 | −0.05040 (19) | 0.35796 (13) | 0.32233 (8) | 0.0132 (3) | |
| C2 | −0.1787 (2) | 0.42658 (14) | 0.32010 (8) | 0.0158 (3) | |
| H2A | −0.2162 | 0.4538 | 0.2799 | 0.019* | |
| C3 | −0.24897 (18) | 0.45302 (14) | 0.37997 (9) | 0.0169 (3) | |
| H3 | −0.3339 | 0.4999 | 0.3803 | 0.020* | |
| C4 | −0.1932 (2) | 0.40991 (13) | 0.43957 (8) | 0.0160 (3) | |
| H4A | −0.2407 | 0.4268 | 0.4798 | 0.019* | |
| C5 | −0.06452 (19) | 0.34082 (13) | 0.43793 (8) | 0.0126 (3) | |
| C6 | 0.00692 (19) | 0.28859 (13) | 0.49932 (8) | 0.0135 (3) | |
| H6 | 0.0363 | 0.3483 | 0.5299 | 0.016* | |
| C7 | −0.1071 (2) | 0.21384 (15) | 0.53495 (9) | 0.0191 (3) | |
| H7A | −0.1304 | 0.1510 | 0.5073 | 0.029* | |
| H7B | −0.1998 | 0.2544 | 0.5438 | 0.029* | |
| H7C | −0.0630 | 0.1888 | 0.5762 | 0.029* | |
| C8 | 0.2787 (2) | 0.24911 (14) | 0.52748 (9) | 0.0162 (4) | |
| H8A | 0.2451 | 0.2360 | 0.5728 | 0.019* | |
| H8B | 0.3086 | 0.3264 | 0.5238 | 0.019* | |
| C9 | 0.4161 (2) | 0.17602 (15) | 0.51239 (8) | 0.0186 (3) | |
| H9A | 0.3824 | 0.0993 | 0.5121 | 0.022* | |
| H9B | 0.4906 | 0.1842 | 0.5481 | 0.022* | |
| C10 | 0.4954 (2) | 0.20050 (14) | 0.44663 (8) | 0.0159 (3) | |
| H10A | 0.5183 | 0.2792 | 0.4450 | 0.019* | |
| H10B | 0.5927 | 0.1611 | 0.4460 | 0.019* | |
| C11 | 0.3963 (2) | 0.04734 (13) | 0.38084 (10) | 0.0193 (3) | |
| H11A | 0.3423 | 0.0199 | 0.4191 | 0.029* | |
| H11B | 0.4979 | 0.0162 | 0.3799 | 0.029* | |
| H11C | 0.3418 | 0.0265 | 0.3411 | 0.029* | |
| C12 | 0.5038 (2) | 0.21068 (15) | 0.32814 (9) | 0.0174 (3) | |
| H12A | 0.6037 | 0.1750 | 0.3311 | 0.021* | |
| H12B | 0.5200 | 0.2897 | 0.3337 | 0.021* | |
| C13 | 0.4389 (2) | 0.19068 (15) | 0.25850 (9) | 0.0177 (3) | |
| H13A | 0.5213 | 0.1989 | 0.2264 | 0.021* | |
| H13B | 0.4023 | 0.1148 | 0.2558 | 0.021* | |
| C14 | 0.3084 (2) | 0.26795 (13) | 0.23923 (8) | 0.0164 (3) | |
| H14A | 0.3422 | 0.3441 | 0.2444 | 0.020* | |
| H14B | 0.2825 | 0.2564 | 0.1927 | 0.020* | |
| C15 | 0.04536 (19) | 0.33025 (14) | 0.26215 (8) | 0.0142 (3) | |
| H15 | 0.0967 | 0.3989 | 0.2484 | 0.017* | |
| C16 | −0.0511 (2) | 0.29131 (16) | 0.20309 (8) | 0.0216 (4) | |
| H16A | 0.0151 | 0.2758 | 0.1660 | 0.032* | |
| H16B | −0.1229 | 0.3483 | 0.1909 | 0.032* | |
| H16C | −0.1061 | 0.2253 | 0.2151 | 0.032* | |
| Cl1 | 0.31018 (5) | 0.52884 (3) | 0.37452 (2) | 0.01419 (7) | |
| O1 | 0.40486 (16) | 0.60559 (10) | 0.41159 (6) | 0.0200 (2) | |
| O2 | 0.15073 (13) | 0.55835 (10) | 0.38070 (7) | 0.0214 (2) | |
| O3 | 0.33372 (15) | 0.41828 (10) | 0.40105 (7) | 0.0221 (3) | |
| O4 | 0.35382 (17) | 0.53023 (11) | 0.30525 (6) | 0.0254 (3) | |
| Cl2 | −0.09242 (5) | 0.01135 (3) | 0.35753 (2) | 0.01541 (8) | |
| O5 | −0.14975 (19) | 0.09033 (13) | 0.30953 (8) | 0.0323 (4) | |
| O6 | 0.05744 (15) | 0.04607 (11) | 0.38012 (8) | 0.0289 (3) | |
| O7 | −0.1936 (2) | 0.00640 (14) | 0.41356 (8) | 0.0373 (4) | |
| O8 | −0.08306 (17) | −0.09626 (11) | 0.32766 (7) | 0.0249 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu1 | 0.01167 (8) | 0.01105 (8) | 0.01101 (8) | 0.00167 (6) | −0.00066 (9) | 0.00045 (6) |
| N1 | 0.0128 (6) | 0.0102 (5) | 0.0133 (6) | −0.0014 (4) | 0.0012 (5) | 0.0002 (5) |
| N2 | 0.0142 (6) | 0.0122 (6) | 0.0130 (6) | 0.0025 (5) | −0.0011 (5) | −0.0005 (5) |
| N3 | 0.0126 (6) | 0.0100 (5) | 0.0173 (6) | −0.0007 (5) | −0.0024 (5) | −0.0011 (5) |
| N4 | 0.0152 (8) | 0.0105 (6) | 0.0144 (6) | 0.0008 (5) | 0.0011 (5) | 0.0002 (4) |
| C1 | 0.0128 (7) | 0.0127 (7) | 0.0141 (7) | −0.0009 (5) | −0.0001 (6) | −0.0001 (6) |
| C2 | 0.0154 (8) | 0.0162 (7) | 0.0156 (7) | 0.0016 (6) | −0.0027 (6) | 0.0016 (6) |
| C3 | 0.0140 (6) | 0.0167 (7) | 0.0201 (7) | 0.0034 (5) | 0.0009 (6) | 0.0001 (7) |
| C4 | 0.0144 (7) | 0.0185 (7) | 0.0151 (7) | 0.0023 (6) | 0.0018 (6) | −0.0011 (6) |
| C5 | 0.0136 (7) | 0.0123 (7) | 0.0121 (7) | −0.0014 (6) | −0.0006 (5) | 0.0005 (5) |
| C6 | 0.0151 (8) | 0.0129 (7) | 0.0124 (7) | 0.0017 (6) | 0.0004 (6) | 0.0001 (6) |
| C7 | 0.0206 (8) | 0.0182 (8) | 0.0185 (8) | 0.0021 (7) | 0.0052 (7) | 0.0038 (6) |
| C8 | 0.0149 (10) | 0.0211 (8) | 0.0126 (7) | −0.0006 (6) | −0.0029 (6) | 0.0000 (6) |
| C9 | 0.0173 (8) | 0.0214 (8) | 0.0170 (8) | 0.0015 (7) | −0.0028 (6) | 0.0033 (6) |
| C10 | 0.0136 (7) | 0.0173 (8) | 0.0167 (7) | 0.0000 (6) | −0.0031 (6) | 0.0001 (6) |
| C11 | 0.0196 (7) | 0.0119 (7) | 0.0264 (8) | 0.0012 (6) | −0.0035 (7) | −0.0011 (7) |
| C12 | 0.0131 (8) | 0.0208 (8) | 0.0182 (8) | −0.0005 (6) | 0.0011 (6) | −0.0015 (6) |
| C13 | 0.0167 (8) | 0.0188 (8) | 0.0176 (8) | 0.0026 (6) | 0.0034 (6) | −0.0024 (6) |
| C14 | 0.0171 (8) | 0.0175 (7) | 0.0147 (7) | −0.0001 (7) | 0.0037 (7) | −0.0007 (5) |
| C15 | 0.0162 (8) | 0.0142 (7) | 0.0122 (7) | 0.0033 (6) | 0.0014 (6) | 0.0020 (5) |
| C16 | 0.0247 (9) | 0.0277 (9) | 0.0124 (7) | 0.0078 (7) | −0.0030 (6) | 0.0000 (7) |
| Cl1 | 0.01690 (16) | 0.01039 (15) | 0.01528 (15) | −0.00062 (12) | 0.00114 (15) | −0.00024 (12) |
| O1 | 0.0203 (6) | 0.0171 (6) | 0.0225 (6) | −0.0045 (5) | −0.0008 (5) | −0.0051 (5) |
| O2 | 0.0157 (5) | 0.0206 (6) | 0.0280 (6) | 0.0019 (4) | −0.0006 (5) | 0.0012 (5) |
| O3 | 0.0252 (7) | 0.0109 (5) | 0.0303 (6) | −0.0007 (5) | −0.0027 (5) | 0.0048 (5) |
| O4 | 0.0359 (8) | 0.0230 (7) | 0.0175 (6) | 0.0010 (6) | 0.0083 (5) | −0.0005 (5) |
| Cl2 | 0.01338 (16) | 0.01372 (16) | 0.01913 (17) | −0.00071 (13) | 0.00050 (15) | −0.00212 (13) |
| O5 | 0.0370 (9) | 0.0249 (7) | 0.0349 (8) | 0.0108 (6) | −0.0117 (6) | 0.0023 (6) |
| O6 | 0.0200 (6) | 0.0197 (6) | 0.0470 (8) | −0.0074 (5) | −0.0096 (7) | 0.0012 (6) |
| O7 | 0.0401 (9) | 0.0351 (8) | 0.0368 (8) | −0.0028 (7) | 0.0230 (8) | −0.0050 (6) |
| O8 | 0.0283 (7) | 0.0176 (6) | 0.0287 (7) | 0.0024 (5) | −0.0031 (6) | −0.0091 (5) |
Geometric parameters (Å, º)
| Cu1—N1 | 1.9316 (13) | C8—H8B | 0.9700 |
| Cu1—N2 | 2.0417 (14) | C8—C9 | 1.515 (3) |
| Cu1—N3 | 2.0108 (13) | C9—H9A | 0.9700 |
| Cu1—N4 | 2.0444 (15) | C9—H9B | 0.9700 |
| Cu1—O3 | 2.4736 (13) | C9—C10 | 1.519 (2) |
| Cu1—O6 | 2.6478 (13) | C10—H10A | 0.9700 |
| N1—C1 | 1.337 (2) | C10—H10B | 0.9700 |
| N1—C5 | 1.329 (2) | C11—H11A | 0.9600 |
| N2—H2 | 0.83 (2) | C11—H11B | 0.9600 |
| N2—C6 | 1.500 (2) | C11—H11C | 0.9600 |
| N2—C8 | 1.485 (2) | C12—H12A | 0.9700 |
| N3—C10 | 1.507 (2) | C12—H12B | 0.9700 |
| N3—C11 | 1.4857 (19) | C12—C13 | 1.525 (2) |
| N3—C12 | 1.496 (2) | C13—H13A | 0.9700 |
| N4—H4 | 0.86 (2) | C13—H13B | 0.9700 |
| N4—C14 | 1.486 (2) | C13—C14 | 1.517 (3) |
| N4—C15 | 1.497 (2) | C14—H14A | 0.9700 |
| C1—C2 | 1.389 (2) | C14—H14B | 0.9700 |
| C1—C15 | 1.506 (2) | C15—H15 | 0.9800 |
| C2—H2A | 0.9300 | C15—C16 | 1.524 (2) |
| C2—C3 | 1.386 (2) | C16—H16A | 0.9600 |
| C3—H3 | 0.9300 | C16—H16B | 0.9600 |
| C3—C4 | 1.390 (2) | C16—H16C | 0.9600 |
| C4—H4A | 0.9300 | Cl1—O1 | 1.4436 (13) |
| C4—C5 | 1.395 (2) | Cl1—O2 | 1.4365 (12) |
| C5—C6 | 1.515 (2) | Cl1—O3 | 1.4498 (12) |
| C6—H6 | 0.9800 | Cl1—O4 | 1.4421 (13) |
| C6—C7 | 1.520 (2) | Cl2—O5 | 1.4423 (15) |
| C7—H7A | 0.9600 | Cl2—O6 | 1.4403 (13) |
| C7—H7B | 0.9600 | Cl2—O7 | 1.4308 (15) |
| C7—H7C | 0.9600 | Cl2—O8 | 1.4317 (13) |
| C8—H8A | 0.9700 | ||
| N1—Cu1—N2 | 82.92 (6) | H8A—C8—H8B | 108.0 |
| N1—Cu1—N3 | 176.39 (5) | C9—C8—H8A | 109.4 |
| N1—Cu1—N4 | 81.79 (6) | C9—C8—H8B | 109.4 |
| N1—Cu1—O3 | 90.40 (5) | C8—C9—H9A | 108.6 |
| N1—Cu1—O6 | 91.30 (5) | C8—C9—H9B | 108.6 |
| N2—Cu1—N4 | 161.21 (6) | C8—C9—C10 | 114.84 (14) |
| N2—Cu1—O3 | 91.20 (5) | H9A—C9—H9B | 107.5 |
| N2—Cu1—O6 | 80.69 (6) | C10—C9—H9A | 108.6 |
| N3—Cu1—N2 | 96.91 (6) | C10—C9—H9B | 108.6 |
| N3—Cu1—N4 | 99.04 (6) | N3—C10—C9 | 116.03 (14) |
| N3—Cu1—O3 | 86.00 (5) | N3—C10—H10A | 108.3 |
| N3—Cu1—O6 | 92.23 (5) | N3—C10—H10B | 108.3 |
| N4—Cu1—O3 | 99.76 (5) | C9—C10—H10A | 108.3 |
| N4—Cu1—O6 | 88.79 (5) | C9—C10—H10B | 108.3 |
| O3—Cu1—O6 | 171.44 (5) | H10A—C10—H10B | 107.4 |
| C1—N1—Cu1 | 119.14 (11) | N3—C11—H11A | 109.5 |
| C5—N1—Cu1 | 118.27 (11) | N3—C11—H11B | 109.5 |
| C5—N1—C1 | 122.07 (14) | N3—C11—H11C | 109.5 |
| Cu1—N2—H2 | 99.0 (15) | H11A—C11—H11B | 109.5 |
| C6—N2—Cu1 | 111.60 (10) | H11A—C11—H11C | 109.5 |
| C6—N2—H2 | 108.8 (16) | H11B—C11—H11C | 109.5 |
| C8—N2—Cu1 | 116.33 (11) | N3—C12—H12A | 108.3 |
| C8—N2—H2 | 108.1 (15) | N3—C12—H12B | 108.3 |
| C8—N2—C6 | 111.95 (13) | N3—C12—C13 | 115.72 (14) |
| C10—N3—Cu1 | 112.38 (10) | H12A—C12—H12B | 107.4 |
| C11—N3—Cu1 | 111.48 (10) | C13—C12—H12A | 108.3 |
| C11—N3—C10 | 108.37 (13) | C13—C12—H12B | 108.3 |
| C11—N3—C12 | 108.83 (13) | C12—C13—H13A | 108.7 |
| C12—N3—Cu1 | 110.52 (10) | C12—C13—H13B | 108.7 |
| C12—N3—C10 | 105.00 (12) | H13A—C13—H13B | 107.6 |
| Cu1—N4—H4 | 101.8 (14) | C14—C13—C12 | 114.27 (14) |
| C14—N4—Cu1 | 117.72 (11) | C14—C13—H13A | 108.7 |
| C14—N4—H4 | 112.3 (14) | C14—C13—H13B | 108.7 |
| C14—N4—C15 | 110.53 (13) | N4—C14—C13 | 112.03 (13) |
| C15—N4—Cu1 | 111.25 (10) | N4—C14—H14A | 109.2 |
| C15—N4—H4 | 101.8 (14) | N4—C14—H14B | 109.2 |
| N1—C1—C2 | 121.18 (15) | C13—C14—H14A | 109.2 |
| N1—C1—C15 | 115.19 (14) | C13—C14—H14B | 109.2 |
| C2—C1—C15 | 123.48 (14) | H14A—C14—H14B | 107.9 |
| C1—C2—H2A | 121.2 | N4—C15—C1 | 110.16 (13) |
| C3—C2—C1 | 117.68 (15) | N4—C15—H15 | 106.9 |
| C3—C2—H2A | 121.2 | N4—C15—C16 | 112.66 (14) |
| C2—C3—H3 | 119.8 | C1—C15—H15 | 106.9 |
| C2—C3—C4 | 120.40 (15) | C1—C15—C16 | 112.84 (14) |
| C4—C3—H3 | 119.8 | C16—C15—H15 | 106.9 |
| C3—C4—H4A | 120.6 | C15—C16—H16A | 109.5 |
| C3—C4—C5 | 118.75 (15) | C15—C16—H16B | 109.5 |
| C5—C4—H4A | 120.6 | C15—C16—H16C | 109.5 |
| N1—C5—C4 | 119.91 (14) | H16A—C16—H16B | 109.5 |
| N1—C5—C6 | 116.31 (14) | H16A—C16—H16C | 109.5 |
| C4—C5—C6 | 123.77 (15) | H16B—C16—H16C | 109.5 |
| N2—C6—C5 | 110.17 (13) | O1—Cl1—O3 | 108.70 (8) |
| N2—C6—H6 | 108.1 | O2—Cl1—O1 | 110.21 (8) |
| N2—C6—C7 | 111.11 (13) | O2—Cl1—O3 | 109.36 (8) |
| C5—C6—H6 | 108.1 | O2—Cl1—O4 | 109.67 (9) |
| C5—C6—C7 | 111.28 (14) | O4—Cl1—O1 | 109.77 (8) |
| C7—C6—H6 | 108.1 | O4—Cl1—O3 | 109.11 (8) |
| C6—C7—H7A | 109.5 | Cl1—O3—Cu1 | 132.04 (8) |
| C6—C7—H7B | 109.5 | O6—Cl2—O5 | 109.19 (9) |
| C6—C7—H7C | 109.5 | O7—Cl2—O5 | 109.90 (10) |
| H7A—C7—H7B | 109.5 | O7—Cl2—O6 | 108.79 (11) |
| H7A—C7—H7C | 109.5 | O7—Cl2—O8 | 109.08 (9) |
| H7B—C7—H7C | 109.5 | O8—Cl2—O5 | 109.81 (9) |
| N2—C8—H8A | 109.4 | O8—Cl2—O6 | 110.05 (9) |
| N2—C8—H8B | 109.4 | Cl2—O6—Cu1 | 132.63 (8) |
| N2—C8—C9 | 111.04 (14) | ||
| Cu1—N1—C1—C2 | −171.12 (12) | C3—C4—C5—N1 | 0.4 (2) |
| Cu1—N1—C1—C15 | 4.49 (19) | C3—C4—C5—C6 | −179.88 (15) |
| Cu1—N1—C5—C4 | 170.68 (12) | C4—C5—C6—N2 | −175.87 (15) |
| Cu1—N1—C5—C6 | −9.09 (18) | C4—C5—C6—C7 | 60.4 (2) |
| Cu1—N2—C6—C5 | 2.44 (16) | C5—N1—C1—C2 | 0.5 (2) |
| Cu1—N2—C6—C7 | 126.24 (12) | C5—N1—C1—C15 | 176.06 (14) |
| Cu1—N2—C8—C9 | −56.21 (16) | C6—N2—C8—C9 | 173.83 (13) |
| Cu1—N3—C10—C9 | 55.43 (16) | C8—N2—C6—C5 | 134.81 (14) |
| Cu1—N3—C12—C13 | −58.20 (16) | C8—N2—C6—C7 | −101.40 (16) |
| Cu1—N4—C14—C13 | 48.28 (16) | C8—C9—C10—N3 | −70.04 (19) |
| Cu1—N4—C15—C1 | −15.39 (16) | C10—N3—C12—C13 | −179.61 (15) |
| Cu1—N4—C15—C16 | −142.35 (12) | C11—N3—C10—C9 | −68.21 (18) |
| N1—C1—C2—C3 | 0.6 (2) | C11—N3—C12—C13 | 64.54 (18) |
| N1—C1—C15—N4 | 7.8 (2) | C12—N3—C10—C9 | 175.62 (15) |
| N1—C1—C15—C16 | 134.62 (15) | C12—C13—C14—N4 | −66.11 (19) |
| N1—C5—C6—N2 | 3.89 (19) | C14—N4—C15—C1 | −148.11 (14) |
| N1—C5—C6—C7 | −119.80 (16) | C14—N4—C15—C16 | 84.93 (17) |
| N2—C8—C9—C10 | 67.86 (19) | C15—N4—C14—C13 | 177.62 (13) |
| N3—C12—C13—C14 | 75.39 (19) | C15—C1—C2—C3 | −174.60 (15) |
| C1—N1—C5—C4 | −1.0 (2) | O1—Cl1—O3—Cu1 | 169.81 (9) |
| C1—N1—C5—C6 | 179.27 (14) | O2—Cl1—O3—Cu1 | 49.44 (13) |
| C1—C2—C3—C4 | −1.2 (2) | O4—Cl1—O3—Cu1 | −70.51 (12) |
| C2—C1—C15—N4 | −176.75 (15) | O5—Cl2—O6—Cu1 | 22.21 (15) |
| C2—C1—C15—C16 | −49.9 (2) | O7—Cl2—O6—Cu1 | −97.73 (14) |
| C2—C3—C4—C5 | 0.7 (3) | O8—Cl2—O6—Cu1 | 142.81 (11) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···O7i | 0.83 (2) | 2.94 (2) | 3.536 (2) | 130.6 (18) |
| N4—H4···O4ii | 0.86 (2) | 2.45 (2) | 3.1619 (19) | 140.2 (18) |
| N4—H4···O5 | 0.86 (2) | 2.77 (2) | 3.423 (2) | 134.1 (17) |
| C2—H2A···O5iii | 0.93 | 2.70 | 3.587 (2) | 161 |
| C3—H3···O1iv | 0.93 | 2.68 | 3.585 (2) | 165 |
| C4—H4A···O1v | 0.93 | 2.64 | 3.518 (2) | 158 |
Symmetry codes: (i) −x, −y, −z+1; (ii) −x+1/2, y−1/2, −z+1/2; (iii) −x−1/2, y+1/2, −z+1/2; (iv) x−1, y, z; (v) −x, −y+1, −z+1.
References
- Autzen, S., Korth, H.-G., Boese, R., Groot, H. & Sustmann, R. (2003). Eur. J. Inorg. Chem. pp. 1401–1410.
- Boros, E., Rybak-Akimova, E., Holland, J. P., Rietz, T., Rotile, N., Blasi, F., Day, H., Latifi, R. & Caravan, P. (2014). Mol. Pharm. 11, 617–629. [DOI] [PMC free article] [PubMed]
- Bruker (2013). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Caira, M. R., Nassimbeni, L. R. & Wooley, P. R. (1975). Acta Cryst. B31, 1334–1338.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Fernandes, A. S., Gaspar, J., Cabral, M. F., Caneiras, C., Guedes, R., Rueff, J., Castro, M., Costa, J. & Oliveira, N. G. (2007). J. Inorg. Biochem. 101, 849–858. [DOI] [PubMed]
- Herrera, A. M., Kalayda, G. V., Disch, J. S., Wikstrom, J. P., Korendovych, I. V., Staples, R. J., Campana, C. F., Nazarenko, A. Y., Haas, T. E. & Rybak-Akimova, E. V. (2003). Dalton Trans. pp. 4482–4492.
- Karn, J. L. & Busch, D. H. (1966). Nature, 211, 160–162.
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Lindoy, L. F., Rambusch, T., Skelton, B. W. & White, A. H. (2001). J. Chem. Soc. Dalton Trans. pp. 1857–1862.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- Organo, V. G., Filatov, A. S., Quartararo, J. S., Friedman, Z. M. & Rybak-Akimova, E. V. (2009). Inorg. Chem. 48, 8456–8468. [DOI] [PubMed]
- Rezaeivala, M. & Keypour, H. (2014). Coord. Chem. Rev. 280, 203–253.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Ye, W., Staples, R. J. & Rybak-Akimova, E. V. (2012). J. Inorg. Biochem. 115, 1–12. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016009701/zl2666sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016009701/zl2666Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016009701/zl2666sup3.tif
CCDC reference: 1485717
Additional supporting information: crystallographic information; 3D view; checkCIF report





