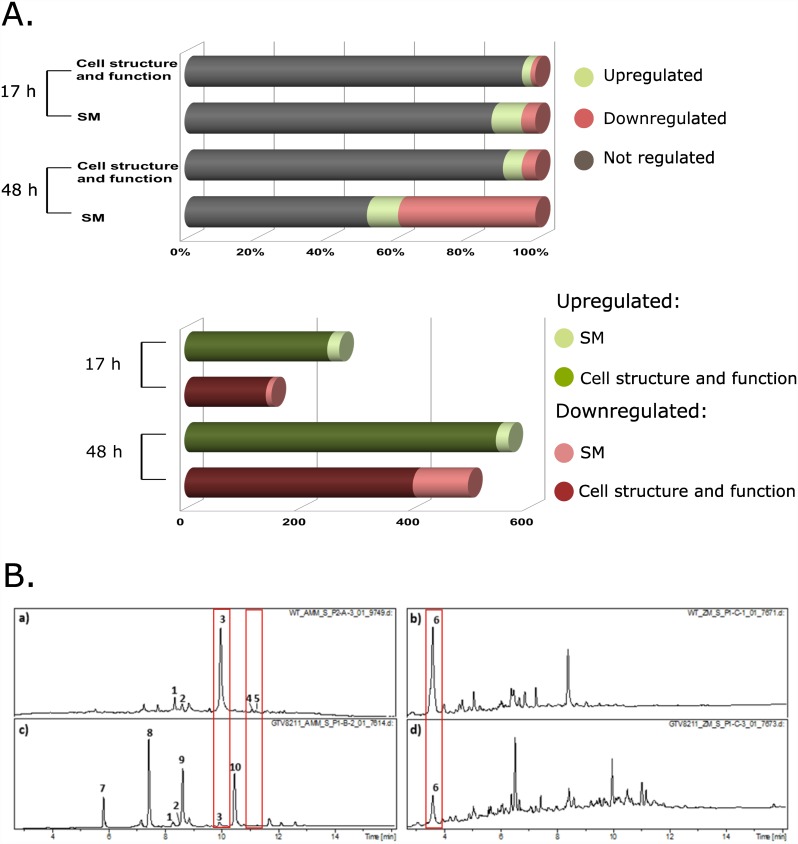
Fig 6. KdmB is required for normal induction of SMB genes.
A. Upper panel: percentage of de-regulated genes in kdmBΔ during PM (17 h) and SM (48 h) with the division into SM cluster genes as annotated in [72] and basic metabolism genes (cell structure and function). Differential expression cut off was set to higher than 4-fold difference (log2 ≥ 2, p<0.05). The lower graph depicts the total number of de-regulated genes in kdmBΔ during PM (17 h) and SM (48 h) cultures,. B. HPLC-chromatograms of supernatant extracts from wild type and kdmBΔ cells growing for 48 hours in conventional AMM or in the special SM-promoting “ZM” medium. a) wild type extract (supernatant, AMM), b) wild type extract (supernatant, ZM), c) kdmBΔ extract (supernatant, AMM), d) kdmBΔ extract (supernatant, ZM). Peaks are assigned to compounds according to standards running in parallel analyses. (1) Austinol, (2) Dehydroaustinol, (3)-Sterigmatocystin, (4) Emericellamide C, (5) Emericellamide D, (6) Orsellinic acid, (7) 2,ω-Dihydroxyemodin, (8) ω-Hydroxyemodin, (9) 2-Hydroxyemodin, (10) Emodin.