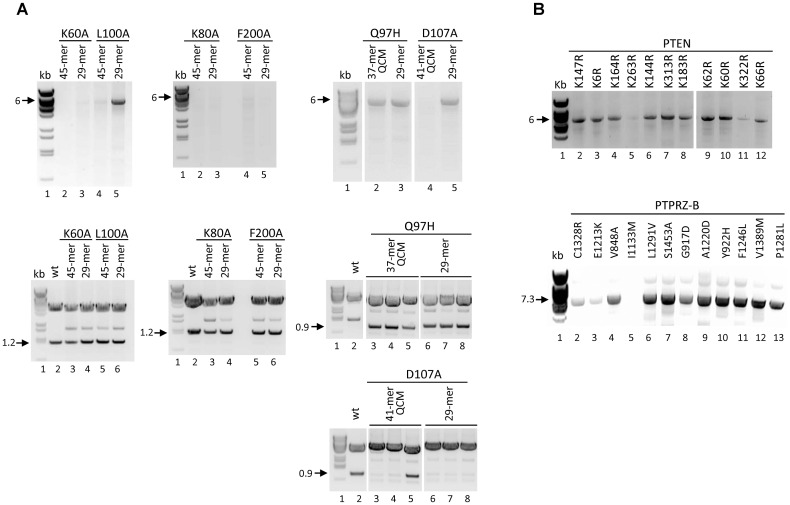
Fig 1. Amino acid substitution mutagenesis using mutagenic primers of different length, Tm, and GC composition.
(A) Mutagenic primer pairs of pre-defined length (45-mer or 29-mer) or as suggested by the web-based QuikChange Primer Design Program (QCM primers) were used to create various PTEN mutations (pRK5-PTEN as template plasmid). Features of the mutagenic primers are listed in Table 1. Upper images show 10 μl of the respective PCR product, or of BstEII-digested λ phage as size marker (kb, lanes 1), following electrophoresis on 1% agarose gels. Lower panels display 1% agarose gel electrophoresis results for purified plasmid DNA (~3 μg) after restriction enzyme digestion. To reveal the 1.2 kbp PTEN insert (mutations K60, L100A, K80A, and F200A) an XbaI/SalI double digestion was used and one sample per SDM reaction is shown. To monitor the mutagenesis efficiency (mutations Q97H and D107A) we used XbaI/SalI or BglII, respectively, and three samples are shown per SDM reaction. Digested wild type pRK5-PTEN (wt, lanes 2) was included as a control. Gels correspond to experiments 1, 3, and 5 listed in Table 1. (B) PCR results using mutagenic (29-nucleotides) primer pairs generating 11 different Lys-to-Arg amino acid substitutions at different PTEN regions (upper panel; pRK5-PTEN as template plasmid), or generating 12 different amino acid substitutions at the intracellular region of PTPRZ-B (lower panel; pENTR-PTPRZ-B as template plasmid). Again, 10 μl of the PCR product was resolved on 1% agarose gels. Corresponding experiments and primer features are listed in Table 2. Molecular sizes of the linearized plasmids (6 and 7.3 kb) or the restriction enzyme-generated DNA fragments, are indicated left of the images in panels A and B.