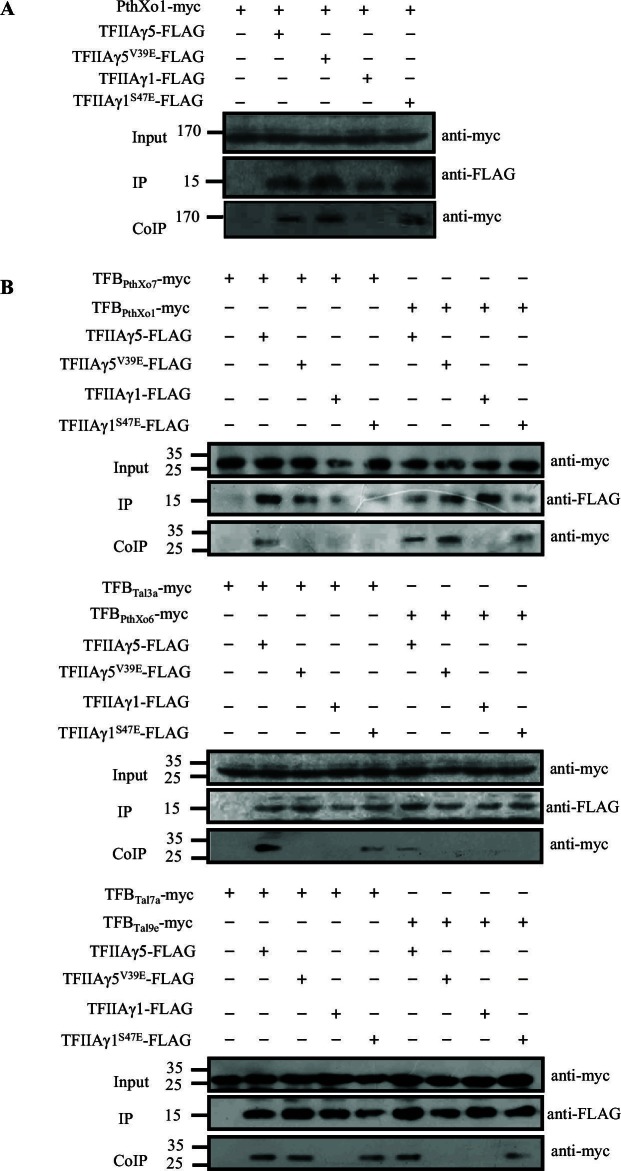
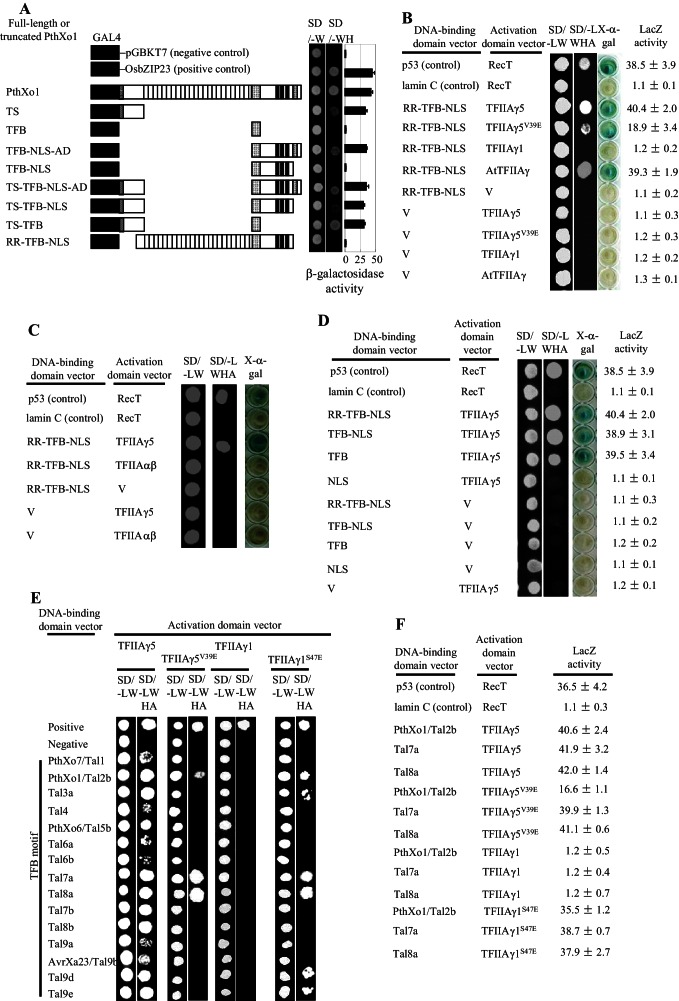
The interactions were assessed by growth of yeast cells on synthetic defined premixes (SD) medium lacking (-) leucine (L), tryptophan (W), histidine (H), and adenine (A). V, empty vector as control. TS, translocation signal; RR, repeat region; TFB, transcription factor binding region; NLS, nuclear localization signal; AD, transcription activation domain. (A) Examination of transactivation activity of different domains and motifs of TALE PthXo1. The full-length and truncated PthXo1 were separately fused to the DNA-binding domain of GAL4, which is a yeast transcription factor, and transformed into yeast. (B) Truncated PthXo1 (RR-TFB-NLS) interacted with rice TFIIAγ5 and TFIIAγ5V39E (mutated TFIIAγ5) and Arabidopsis AtTFIIAγ analysed by yeast two-hybrid (Y2H) assay. (C) Truncated PthXo1 did not interact with rice basal transcription factor TFIIAαβ analysed by Y2H assay. (D) The TFB region of PthXo1 was required for the interaction with TFIIAγ5 analysed by Y2H assay. (E) The TFB regions of TALEs differentially interacted with rice TFIIAγs. The TFB regions of all the 15 TALEs from Xoo strain PXO99 interacted with rice TFIIAγ5, and the TFB regions of some of the 15 TALEs interacted with the mutated TFIIAγs from rice (TFIIAγ5V39E and TFIIAγ1S47E) analysed by Y2H assay. The Tal6b is a putative non-functional TALE. (F) The TFB regions of TALEs interacted with rice TFIIAγs with different strength based on the analysis of LacZ activity.