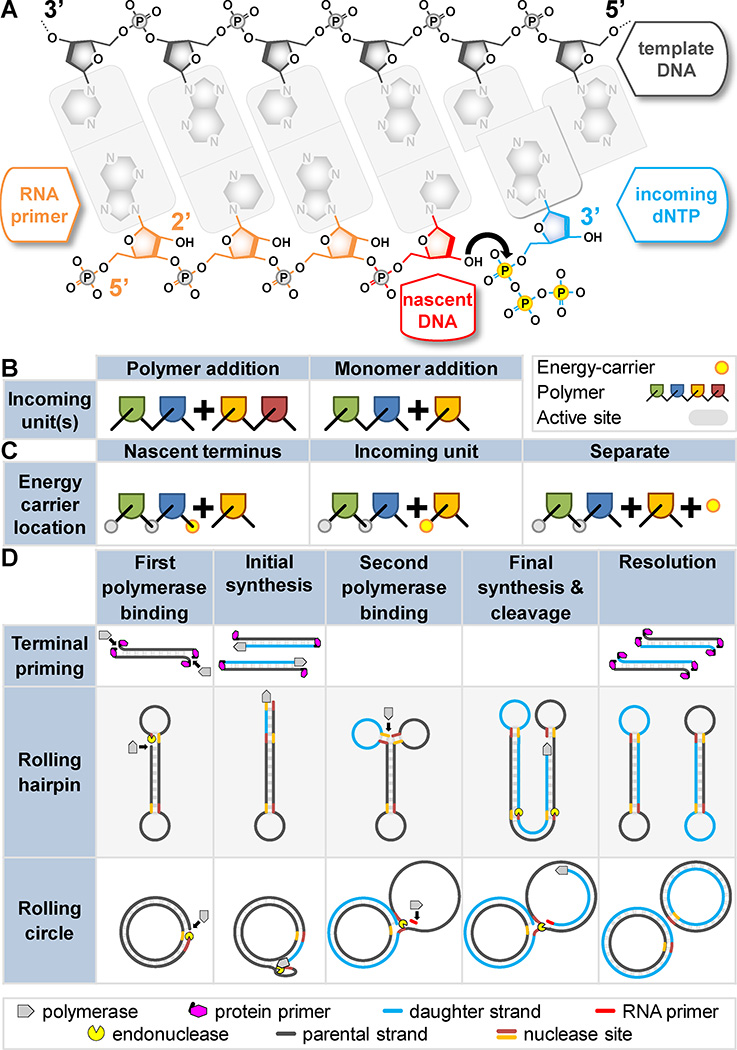
Figure 1. Why DNA replication is asymmetric and how simple replicators cope.
A) Termini of each DNA strand are named according to the hydroxyl groups on the deoxyribose sugar: 5’ and 3’ (labeled). DNA polymerases synthesize DNA via nucleophilic attack (black arrow) by the 3’ hydroxyl of the growing strand (the primer, red) on the α-phosphate of the incoming nucleotide triphosphate (blue). Chemical energy carried by the triphosphate (yellow) drives the reaction. RNA (orange) can also serve as a primer. RNA has a 2’ hydroxyl (labeled) that is absent in DNA. B–D) Genome synthesis could theoretically proceed by many modes. Arguments to which these panels refer hold for any hypothetical information-carrying polymer, thus polymer and terminal identities are herein deliberately ambiguous. Non-replicative polymerases and other enzymes show that alternate chemistries are possible, but replicases are limited: B) monomers (not polymers) are added in each synthesis step; C) the energy carrier is a moiety of the incoming monomer (not the growing strand or another reaction component); D) The leading/lagging asymmetry is avoidable. Terminal priming and rolling hairpin replication, sufficient for small linear genomes, require either a terminal-binding protein (magenta), or terminal hairpins (exposed single-strand), respectively. Rolling circle replication, sufficient for small circular genomes, requires RNA priming (red) on the second strand. Note: a stepwise nature is exaggerated for clarity; second strand synthesis is often simultaneous.