Fig. 5.
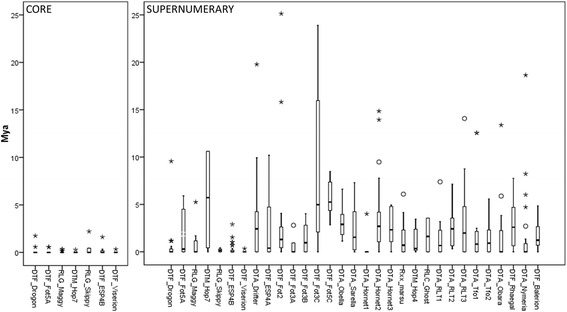
Divergence estimation of intact (not RIPped) TE copies on the core (left) and supernumerary (right) genomes. Copies were aligned and branch lengths extracted from a maximum-likelihood phylogenetic tree. Branch lengths were used to calculate divergence times with a fixed substitution rate (1.05 * 10-9 substitutions per site per year [66]). Y axis scale was cut off at 25 Mya, but for the supernumerary genome many outliers are above this value. Additional file 19 shows the boxplot with outliers for the supernumerary genome. The boxes for every TE show the lower and upper quartile of the divergence estimates and the median (thick line within the boxes). The whiskers represent the minimum and maximum values. Circles and asterisks are outliers and extreme values which fall respectively outside of one-and-a-half additional box lengths and three additional box lengths counted from the upper quartile limit
