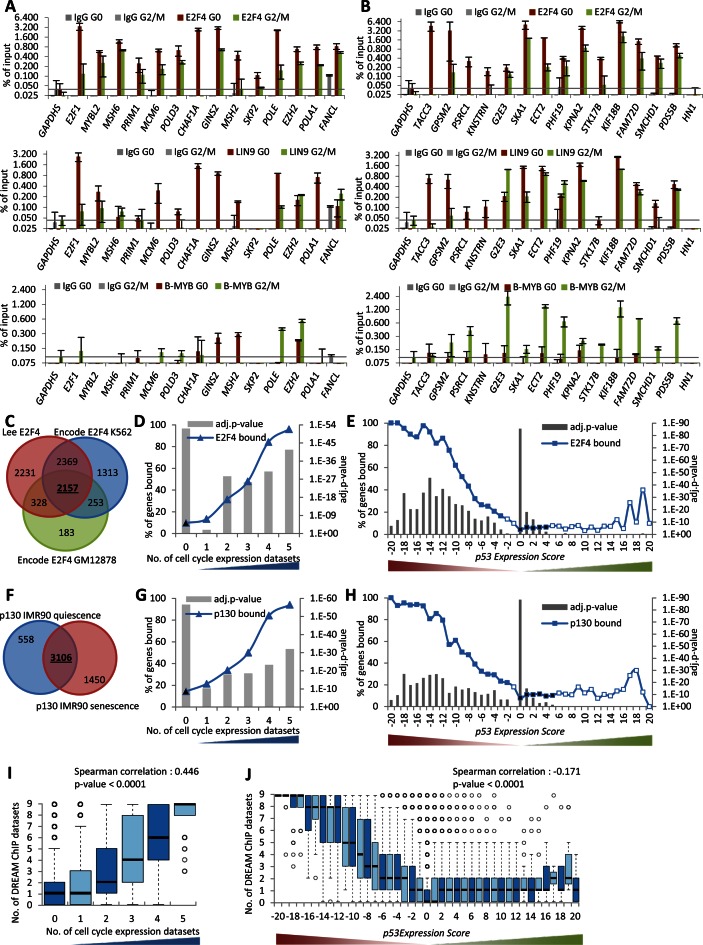
Figure 5.
Prediction and validation of candidate DREAM target genes. (A and B) Protein binding to promoters of the indicated genes was tested by ChIP in serum starved (G0) and 22 h re-stimulated (G2/M) T98G cells. The E2F1 and MYBL2 promoters served as a positive control for DREAM binding; the GAPDHS promoter was used as a negative control. One representative experiment with three technical replicates (n = 3) is displayed. Venn diagram of overlaps between genes identified as bound in the (C) E2F4 or (F) p130 ChIP-seq datasets. The number of common (D) E2F4 or (G) p130 bound genes is compared to the number of datasets that identify a gene as being a CC gene. The number of common (E) E2F4 or (H) p130 bound genes in the 41 p53 Expression Score groups. A two-sided Fisher's exact test was employed to test for significant over- and under-representation of gene sets and P-values were adjusted for multiple testing using Bonferroni correction. Colored and black data points are significantly over- and under-represented, respectively (adj. P-value ≤ 0.05). White data points are not significantly different. (I) Boxplot displaying the number of datasets that find a gene to be targeted by DREAM compared to the number of datasets that identify a gene as CC regulated. (J) Boxplot displaying the number of datasets that find a gene to be targeted by DREAM in the 41 p53 Expression Score groups. Correlation coefficient and two-tailed P-value was calculated using GraphPad Prism version 6.00.