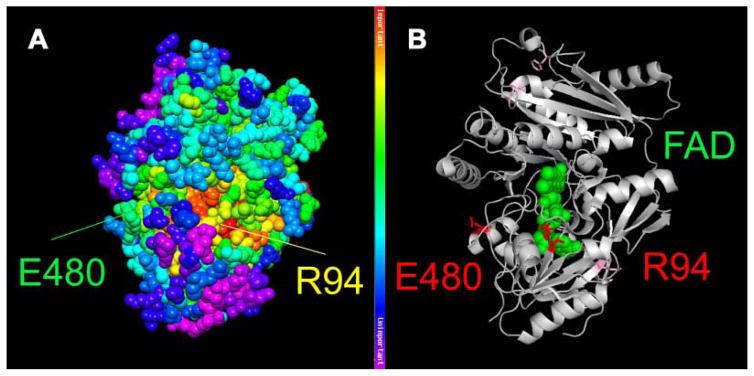
FIG. 3.
Structural model of DHCR24. Mapping of the patient’s mutations (p.R94H and p.E480K) onto the closest homologue for which structural data is available (cytokinin oxidase/dehydrogenase; CKX) from Arabidopsis thaliana (PDB id: 2exr). A: Mapping provides information about evolutionary importance of respective amino acids, with red being most highly conserved and purple being least conserved. B: Depiction of the two affected amino acids in red and mutations of previously reported patients are shown as light purple sticks. An FAD molecule is represented as green balls, illustrating that the p.R94H substitution affects the FAD binding domain of the protein. The figure was generated by using PyMOL (DeLano Scientific LLC, Palo Alto, CA) and the Protein Data Bank file: 2exr.