Abstract
Increased APOBEC3B mRNA levels are associated with a hypermutator phenotype and poor prognosis in ER-positive breast cancer patients. In addition, a 29.5 kb deletion polymorphism of APOBEC3B, resulting in an APOBEC3A-B hybrid transcript, has been associated with an increased breast cancer risk and the hypermutator phenotype. Here we evaluated whether the APOBEC3B deletion polymorphism also associates with clinical outcome of breast cancer. Copy number analysis was performed by quantitative PCR (qPCR) in primary tumors of 1,756 Dutch breast cancer patients. The APOBEC3B deletion was found in 187 patients of whom 16 carried a two-copy deletion and 171 carried a one-copy deletion. The prognostic value of the APOBEC3B deletion for the natural course of the disease was evaluated among 1,076 lymph-node negative (LNN) patients who did not receive adjuvant systemic treatment. No association was found between APOBEC3B copy number values and the length of metastasis-free survival (MFS; hazard ratio (HR) = 1.00, 95% confidence interval (CI) = 0.90–1.11, P = 0.96). Subgroup analysis by ER status also did not reveal an association between APOBEC3B copy number values and the length of MFS. The predictive value of the APOBEC3B deletion was assessed among 329 ER-positive breast cancer patients who received tamoxifen as the first-line therapy for recurrent disease and 226 breast cancer patients who received first-line chemotherapy for recurrent disease. No association between APOBEC3B copy number values and the overall response rate (ORR) to either tamoxifen (odds ratio (OR) = 0.88, 95% CI = 0.69–1.13, P = 0.31) or chemotherapy (OR = 0.97, 95% CI = 0.71–1.33, P = 0.87) was found. Thus, in contrast to APOBEC3B mRNA levels, the APOBEC3B deletion polymorphism has neither a prognostic nor a predictive value for breast cancer patients. Although a correlation exists between APOBEC3B copy number and mRNA expression, it is relatively weak. This suggests that other mechanisms exist that may affect and therefore determine the prognostic value of APOBEC3B mRNA levels.
Introduction
Breast cancer, like most cancer types, is a heterogeneous disease. The heterogeneous nature of breast cancer, however, provides challenges for identifying appropriate markers for disease susceptibility and progression, as well as treatment selection. Accordingly, transcriptional profiling has identified five molecular subtypes of breast cancer, which differ in prognosis, efficacy of treatment and preferred site of metastasis [1–5]. More recently, the catalogues of mutations across human cancers have provided us insight into the mutational processes that drive tumorigenesis [6,7]. For breast cancer, five distinct mutational signatures have been defined that contribute in varying degree to the final mutational catalogue of a breast tumor [7]. One of the most pronounced mutational processes impacting breast tumorigenesis is driven by the AID/APOBEC family of cytidine deaminases and gives rise to C>T and C>G substitutions at TpCpN nucleotides. Moreover, this mutational process associates with regional somatic hypermutation or kataegis [6–8].
The APOBEC3 gene cluster is located on chromosome 22q13.1-q13.2 and harbors seven APOBEC3 genes that have evolved in primates (i.e. APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3D, APOBEC3F, APOBEC3G and APOBEC3H) [9]. APOBEC3s play a role in intracellular defense through restriction of retroviral infections, but also of infections from the cancer-associated hepatitis B virus, the human papilloma virus and human T-lymphotropic virus [10]. Moreover, APOBEC3A, APOBEC3B, APOBEC3C and APOBEC3F are also able to inhibit LINE1 retrotransposition [11,12]. Besides its role in innate immunity, APOBEC3B has recently been identified as an endogenous source of mutation in breast cancer [13]. APOBEC3B mRNA expression was found to be upregulated in most breast cancers and tumors expressing high levels of APOBEC3B had a 2-fold increase in mutations compared with tumors expressing low APOBEC3B levels. This suggests that APOBEC3B, at least in part, underlies the APOBEC-driven mutational process in breast cancer, but also in other cancers [13,14]. In line with these findings, high levels of APOBEC3B mRNA were associated with a shorter disease-free survival in ER-positive, LNN, systemically untreated patients, as well as with earlier recurrence in luminal subtype patients and with a more aggressive phenotype in Japanese breast cancers [15–17]. Moreover, APOBEC3B expression has been reported to be associated with a strong enrichment of mitotic and cell cycle-related genes [16].
A 29.5 kb germline deletion between the fifth exon of APOBEC3A and the eighth exon of APOBEC3B has been identified that essentially removes the complete APOBEC3B coding region from the genome and generates a fusion transcript of APOBEC3A with the 3’untranslated region (UTR) of APOBEC3B [18]. With a worldwide frequency of 22.5%, the frequency of the germline APOBEC3B deletion variant varies widely among the different ethnic groups, ranging from being rare in African and European populations (i.e. 0.9% and 6%, respectively) to being common in Asian and American populations (i.e. 36.9% and 57.7%, respectively) [18]. Through a genome-wide association study of copy number variation, Long et al. found that the APOBEC3B deletion variant was associated with an increased risk to develop breast cancer in Chinese women [19]. This finding was replicated among European [20] and Southeast Iranian women [21], but not among Swedish women [22]. Interestingly, carriers of the APOBEC3B deletion were shown to have a greater APOBEC3A mRNA stability resulting in higher APOBEC3A levels, increased activity of APOBEC-driven mutational processes and more severe DNA damage [23,24]. However, the APOBEC3B deletion polymorphism was not associated with the survival of breast cancer patients [16,22]. Thus, despite that APOBEC3B overexpression and the APOBEC3B deletion variant both result in a hypermutation phenotype, there seems to be a difference between the two mechanisms and their association with clinical outcome.
To investigate the relation between the APOBEC3B deletion polymorphism and clinical outcome of 1,756 Dutch breast cancer patients, we explored four different clinical cohorts: LNN patients who did not receive any adjuvant treatment, lymph node positive (LNP) patients who did receive adjuvant systemic treatment, hormone-naive ER-positive patients who received tamoxifen as first-line therapy for recurrent disease and patients that received first-line chemotherapy for recurrent disease. Furthermore, to investigate the relation between the clinical outcomes based on the APOBEC3B deletion polymorphism versus APOBEC3B overexpression, we investigated the correlation between APOBEC3B copy number and APOBEC3B mRNA expression.
Materials and Methods
Study population
The retrospective study cohort consisted of 1,756 breast cancer patients who underwent surgery for an invasive primary breast cancer between 1978 and 2001. Inclusion criteria were no neoadjuvant treatment, no experience of a previous other cancer (except for basal cell carcinoma or stage Ia/Ib cervical cancer), a minimum of 100 mg of freshly frozen primary tumor tissue available for downstream DNA isolation and DNA available from tissue with a tumor cell nuclei percentage ≥ 30%. Cytosolic ER and PR levels were determined by ligand binding assay or enzyme immunoassay [25,26]. ER and/or PR positivity was defined by ≥10 fmol/mg cytosolic protein and ERBB2 overexpression was defined by a reverse transcriptase qPCR expression level ≥18 [27]. In total, 796 patients underwent breast-conserving lumpectomy and 960 patients underwent modified radical mastectomy. In addition, 215 patients received adjuvant hormonal therapy, 308 patients received adjuvant chemotherapy and 6 patients received both adjuvant hormonal therapy and chemotherapy. There were 1,713 M0 patients and 43 M1 patients. The median age at the time of the primary surgery was 54 years, while the median age at the start of first-line treatment was 55 years. The clinicopathological variables of the patients are shown in Table 1.
Table 1. Association of APOBEC3B copy number status with clinicopathological variables in 1,756 primary breast cancers.
| Variables | Deleted | Balanced | Amplified | P-value | |||
|---|---|---|---|---|---|---|---|
| Total number | 187 | 1260 | 309 | ||||
| Age (in years) | 0.54 | ||||||
| ≤ 40 | 23 | (12.3%) | 160 | (12.7%) | 48 | (15.5%) | |
| 41–55 | 82 | (43.9%) | 498 | (39.5%) | 116 | (37.5%) | |
| 56–70 | 62 | (33.2%) | 412 | (32.7%) | 99 | (32.0%) | |
| >70 | 20 | (10.7%) | 190 | (15.1%) | 46 | (14.9%) | |
| Menopausal status | 0.31 | ||||||
| Premenopausal | 89 | (47.6%) | 554 | (44.0%) | 149 | (48.2%) | |
| Postmenopausal | 98 | (52.4%) | 706 | (56.0%) | 160 | (51.8%) | |
| Tumor size | 1.00 | ||||||
| pT1 | 71 | (38.0%) | 469 | (37.2%) | 115 | (37.2%) | |
| pT2 + Unknown | 96 | (51.3%) | 661 | (52.5%) | 163 | (52.8%) | |
| pT3 + pT4 | 20 | (10.7%) | 130 | (10.3%) | 31 | (10.0%) | |
| Nodal status | 0.041 | ||||||
| N0 | 117 | (63.6%) | 793 | (63.5%) | 173 | (56.5%) | |
| N1-3 | 23 | (12.5%) | 212 | (17.0%) | 67 | (21.9%) | |
| N>3 | 44 | (23.9%) | 244 | (19.5%) | 66 | (21.6%) | |
| Tumor grade | 0.052 | ||||||
| Good/Moderate | 34 | (23.8%) | 193 | (21.9%) | 66 | (29.6%) | |
| Poor | 109 | (76.2%) | 689 | (78.1%) | 157 | (70.4%) | |
| Tumor histology | 0.29 | ||||||
| IDC | 129 | (83.2%) | 833 | (80.6%) | 196 | (78.4%) | |
| ILC | 10 | (6.5%) | 119 | (11.5%) | 30 | (12.0%) | |
| Other | 16 | (10.3%) | 82 | (7.9%) | 24 | (9.6%) | |
| ER status | 0.24 | ||||||
| Positive | 130 | (70.3%) | 951 | (75.8%) | 226 | (73.9%) | |
| Negative | 55 | (29.7%) | 303 | (24.2%) | 80 | (26.1%) | |
| PR status | 0.99 | ||||||
| Positive | 114 | (65.5%) | 779 | (65.8%) | 187 | (65.6%) | |
| Negative | 60 | (34.5%) | 404 | (34.2%) | 98 | (34.4%) | |
| HER2 status | 0.59 | ||||||
| Positive | 18 | (12.6%) | 150 | (15.7%) | 39 | (16.2%) | |
| Negative | 125 | (87.4%) | 807 | (84.3%) | 201 | (83.8%) | |
| Adjuvant systemic therapy | 0.17 | ||||||
| None | 133 | (73.5%) | 856 | (69.9%) | 191 | (63.0%) | |
| Chemotherapy | 26 | (14.4%) | 219 | (17.9%) | 63 | (20.8%) | |
| Hormonal therapy | 22 | (12.2%) | 146 | (11.9%) | 47 | (15.5%) | |
| Both | 0 | (0%) | 4 | (0.3%) | 2 | (0.7%) | |
Note: For nodal status, tumor grade, tumor histology, ER, PR and HER2 status and adjuvant systemic therapy the number of patients do not add up to 1,756 because there were missing values for these variables. In addition, for the adjuvant systemic treatment variable: some patients were not eligible for adjuvant treatment because they were had distant metastasis at the time of primary tumor diagnosis.
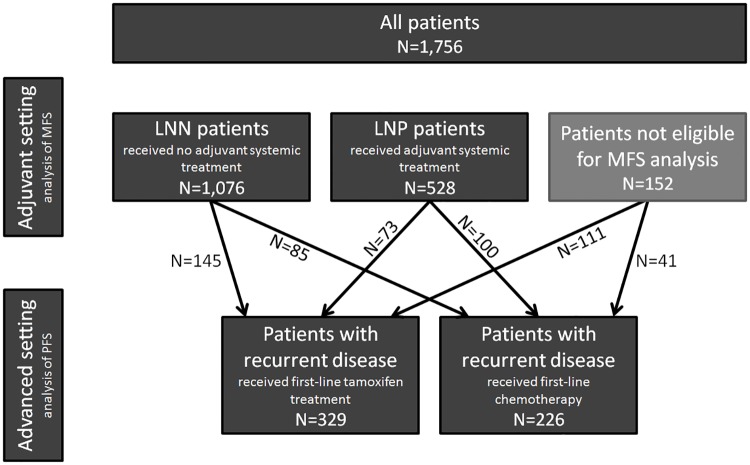
The total study cohort consists of four specific studies that were grouped together: 1) 1,076 LNN patients who did not receive any adjuvant systemic treatment, 2) 528 LNP patients who received adjuvant systemic treatment, 3) 329 hormone-naive ER-positive patients who received first-line tamoxifen therapy for recurrent disease and 4) 226 patients who received first-line chemotherapy for recurrent disease as detailed in Fig 1. We included all eligible patients fulfilling the study criteria and the general inclusion criteria specified above. As a consequence 403 patients were included in two studies (i.e. one study in the adjuvant setting and one study in the advanced setting). The total study population, however, cannot be considered a pure consecutive series, since systemically treated LNN patients were not included. The reason for this is that we especially wished to study the association of APOBEC3B copy number with the natural course of the disease in LNN patients, not potentially confounded by adjuvant systemic therapy.
Fig 1. Schematic overview of study cohort.
In total, this retrospective study consists of 1,756 primary breast cancers from patients who underwent surgery between 1978 and 2001. Inclusion criteria are specified in the Materials and Methods section. In the adjuvant setting, there were 1,076 lymph node negative (LNN) patients who did not receive adjuvant systemic treatment and 528 lymph node positive (LNP) patients who received adjuvant systemic treatment for the analysis of MFS. In the advanced setting, a group of 329 hormone-naive patients with ER-positive breast cancer received first-line tamoxifen for recurrent disease. Of these, 145 patients came from the LNN patients group and 73 came from the LNP patients group. The remaining 111 patients in this group did not qualify for MFS analysis (i.e. 82 patients did not fulfill LNN or LNP study eligibility criteria and 29 patients already presented with metastasis at the time of diagnosis). Furthermore, a group of 226 patients received first-line chemotherapy for recurrent disease. Of these, 85 patients came from the LNN patients group and 100 came from the LNP patients group. The remaining 41 patients in this group did not qualify for MFS analysis (i.e. 27 patients did not fulfill LNN or LNP study eligibility criteria and 14 patients already presented with metastasis at the time of diagnosis).
Routine postsurgical follow-up and the definition of time to metastasis for LNN and LNP patients were as described previously [28]. The median follow-up for the 1,604 LNN and LNP patients included in the prognostic studies for the analysis of metastasis-free survival (MFS) was 114 months (range 10–354 months). Of these 1,604 patients, 686 (42.8%) had developed a distant metastasis and 691 (43.1%) of the 1,604 patients died during follow up. More specifically, 602 (37.5%) patients died after disease recurrence, whereas 89 (5.5%) patients died without evidence of disease recurrence at last follow-up. These 89 patients were censored at the date of last follow-up in the analysis of MFS.
Criteria for follow up and response to tamoxifen therapy were defined by standard International Union Against Cancer (Geneva, Switzerland) criteria of tumor response [29]. Complete and partial remission (together objective response) was observed in 11 and 48 patients, respectively, whereas 79 patients had progressive disease. From the patients with stable disease, 171 had no change for > 6 months, whereas 20 patients had no change for ≤ 6 months. According to the advice of the European Organization for Research and Treatment of Cancer [30], we defined overall response as complete and partial remission including stable disease > 6 months. As a result, 230 patients were classified as responders to tamoxifen and 99 patients showed no response to tamoxifen. The median follow up of patients after start of tamoxifen therapy was 49 months (range: 4–176 months). At the end of the follow up, 304 (92.4%) patients had developed tumor progression and were counted as events in the analysis of progression-free survival (PFS) and 264 patients had died.
Criteria for follow up and response to chemotherapy were similar to those for tamoxifen therapy with the exception that not all patients were chemotherapy-naïve. In fact, 45 out of 226 patients had received adjuvant chemotherapy. Of those, 33 patients received cyclophosphamide/methotrexate/5-fluorouracil (CMF), 11 patients received anthracycline-based chemotherapy and 1 patient received both. Complete and partial remission was observed in 12 and 69 patients, respectively, whereas 66 patients had progressive disease. From the 77 patients with stable disease, 54 had no change for > 6 courses, whereas 23 patients had no change for ≤ 6 courses. We defined overall response as complete and partial remission including stable disease >6 courses. As a result, 135 patients were classified as responders to chemotherapy and 89 patients showed no response to chemotherapy. For two patients the type of response was ambiguous. The median follow up of patients after start of chemotherapy was 30 months (range: 4–153 months). Fifty-two patients received consolidation therapy after chemotherapy and PFS was right censored at two months after the start of consolidation therapy. At this time point, 138 patients had developed tumor progression and were counted as events in the analysis of PFS. At the end of follow up, 210 patients had died.
This study was approved by the Medical Ethical Committee of the Erasmus Medical Center Rotterdam, the Netherlands (MEC 02.953). As this is a retrospective study using remaining material from surgical resection of the patient’s primary tumor, obtaining informed consent from the patient was not required provided patient records were anonymized and de-identified prior to analysis. Herewith, we adhered to the Code of Conduct of the Federation of Medical Scientific Societies in the Netherlands (http://www.fmwv.nl). Results are reported in accordance with the REMARK criteria on clinical reporting [31].
Copy number analysis
Copy number analysis for APOBEC3B was performed on genomic DNA isolated from fresh-frozen primary tumor sections by qPCR on a Mx3000/3005P machine (Agilent Technologies, Santa Clara, CA). Briefly, genomic DNA was isolated from two to ten 30 μm cryostat sections (5–20 mg) with the NucleoSpin Tissue kit (Macherey–Nagel, Düren, Germany) according the protocol provided by the manufacturer. DNA quantity and quality was assessed by Nanodrop and the Quant-iT PicoGreen dsDNA HS Assay Kit (Thermo Scientific, Waltham, MA). Next, 0.5X Taqman Copy Number Assay for the APOBEC3B gene (Hs04504055_cn; Thermo Scientific), 0.5X Taqman Copy Number Reference Assay (i.e. for the RNase P gene; Thermo Scientific) and 0.5X ABsolute qPCR Mix, low ROX (Thermo Scientific) were added to 20 ng of genomic DNA in a final volume of 17 μl. Cycling conditions were: 1 cycle of 15 minutes at 95°C and 45 cycles of 15 seconds at 92°C and 1 minute at 60°C. The MxPro qPCR software v4.10 (Agilent) was used to calculate the cycle threshold (Ct) values. Relative quantification analysis was performed within the CopyCaller v2.0 software (Thermo Scientific). For this, the ΔCt value was calculated for each sample by subtracting the Ct value for the target gene (i.e. APOBEC3B) from the Ct value of the reference gene (i.e. RNase P). In the case of no Ct value for APOBEC3B after 45 cycles, while the RNase P gene was successfully amplified within 32 cycles for that sample, the ΔCt value could not be quantified and the sample was designated to have a two-copy deletion of the APOBEC3B gene. For samples where the ΔCt was quantified, ΔCt values were converted to calculated copy number values by the CopyCaller software as detailed in S1 Fig. Then, samples with calculated copy numbers ≤ 0.2 were called as two-copy deletion of the APOBEC3B gene, samples with calculated copy numbers >0.20 and ≤ 1.41 as one-copy deletion of the APOBEC3B gene, samples with calculated copy numbers > 1.41 and ≤ 3.44 as no copy number change or balanced and samples with calculated copy numbers > 3.44 as amplified for the APOBEC3B gene. Furthermore, 325 samples were measured in duplicate distributed over the 96-well sample plates and each 96-well plate included genomic DNA from breast cancer cell line OCUB-F as a control since this cell line has a two-copy deletion of APOBEC3B.
Expression analysis
APOBEC3B mRNA expression analysis has been performed previously for 1,491 breast cancer patients [15]. Extraction of RNA, synthesis of cDNA, reverse transcriptase quantitative PCR and quantification of transcripts was also described before [32]. Out of the 1,756 patients for whom we performed APOBEC3B copy number analysis in the current study, APOBEC3B mRNA expression data was available for 1,132 patients.
Statistical analyses
Because the number of patients carrying a two-copy deletion of APOBEC3B was small (N = 16, 0.91%), we grouped patients with one-copy and two-copy deletions together. A χ2 or Fisher’s exact test (when the expected frequency was ≤ 5 in any of the groups) was used to evaluate the association between the APOBEC3B copy number status (i.e. deleted, balanced or amplified) and the clinicopathological variables. To assess the association between the APOBEC3B copy number status and the ORR to either first-line tamoxifen treatment or first-line chemotherapy, we used a logistic regression model to calculate ORs and their 95% CIs. For visualization purposes, we performed survival analysis by the Kaplan-Meier method. The difference between survival curves for patients with either a deletion, no copy number change, or an amplification was calculated using the 3-sample logrank test. In addition, univariate Cox proportional hazards regression models including continuous calculated copy number values as covariate were performed to assess the association between APOBEC3B copy number and survival times (i.e. MFS in the adjuvant setting and PFS in the advanced setting). Finally, the correlation between APOBEC3B copy number and APOBEC3B expression was evaluated using Spearman’s rank correlation. All P-values were two-sided and P-values < 0.05 were considered to be statistically significant. Analyses were performed using R, version 3.2.3, except for the power and sample size calculations shown in the Discussion. For these, we used the stpower cox tool in Stata version 13.1 (StataCorp, College Station, TX) assuming an alpha of 0.05, a beta of 0.2 (i.e. only in sample size calculations) and similar covariate standard deviations, allele frequencies, event probabilities and sample sizes (i.e. only in power calculations) as observed in the current study.
Results
We have performed APOBEC3B copy number analyses among 1,756 primary breast cancers and found no copy number change among 1,260 breast cancers. A two-copy deletion was identified in 16 (0.91%) breast cancers, whereas 171 (9.74%) breast cancers had a one-copy deletion. In addition, we detected an amplified APOBEC3B gene locus in 309 (17.26%) breast cancers. The minor allele frequency of the APOBEC3B deletion was 5.8% (203/3512) which is similar to the expected frequency in European population (i.e 6.5%) [18].
Next, we evaluated the association between the APOBEC3B copy number status and the clinicopathological variables of the 1,756 breast cancer patients. We found that APOBEC3B copy number status was significantly associated with nodal status (P = 0.041), but not with age, menopausal status, tumor size, tumor grade, tumor histology, ER, PR and HER2 status, and adjuvant systemic therapy (Table 1). Despite the significant association between APOBEC3B copy number status and nodal status, however, no meaningful trend was observed (Table 1).
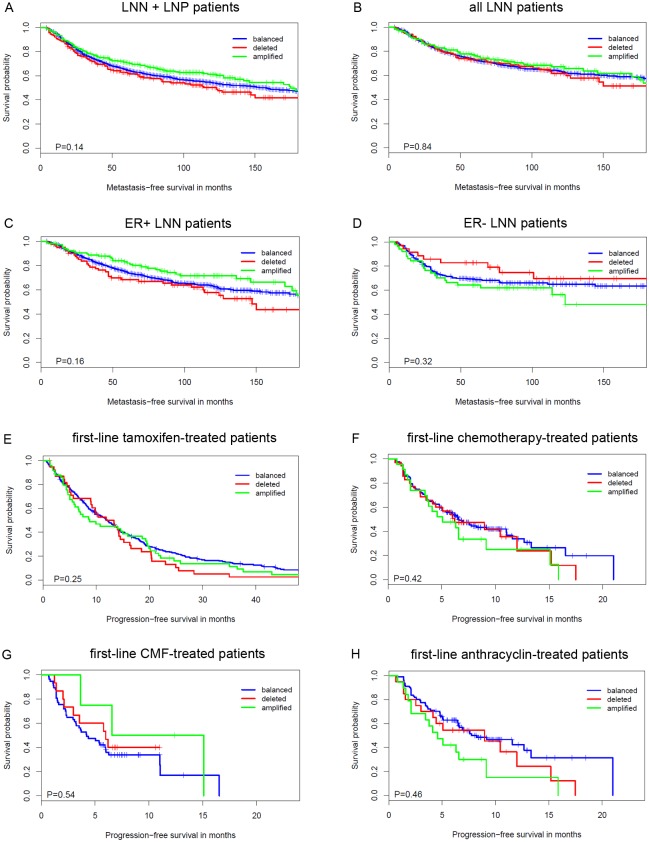
To assess whether APOBEC3B copy numbers associate with clinical outcome in the adjuvant setting, we performed Kaplan-Meier survival analysis and Cox regression analysis in 1,604 LNN and LNP patients. No association between APOBEC3B copy number status and the length of MFS was found (P = 0.14; Fig 2A). Moreover, calculated copy number values were also not associated with the length of MFS (HR = 0.95, 95% CI = 0.88–1.02, P = 0.17; Table 2). Because all LNP patients were treated with adjuvant systemic therapy and we specifically wanted to evaluate the prognostic value of APOBEC3B copy number, we repeated these analyses in the cohort of 1,076 LNN patients that had not received any adjuvant systemic treatment. Again, we found no association between APOBEC3B copy number status and the length of MFS (P = 0.84; Fig 2B), nor did we find an association between APOBEC3B calculated copy number values and the length of MFS (HR = 1.00, 95% CI = 0.90–1.11, P = 0.96; Table 2). Also when we performed subgroup analysis in 769 ER-positive or 300 ER-negative untreated LNN patients (i.e. ER status was not available for 7 patients), APOBEC3B copy numbers did not appear to have any prognostic value (Fig 2C and 2D, Table 2).
Fig 2. Kaplan-Meier survival analysis as a function of APOBEC3B copy number status.
(A) In 1,604 patients of the lymph node negative (LNN) and lymph node positive (LNP) cohort combined. (B) In 1,076 LNN patients who did not receive any adjuvant systemic treatment. (C) In 769 ER-positive LNN patients who did not receive any adjuvant systemic treatment. (D) In 300 ER-negative LNN patients who did not receive any adjuvant systemic treatment. (E) In 329 ER-positive breast cancer patients who received first-line tamoxifen for recurrent disease. (F) In 226 breast cancer patients who received first-line chemotherapy for recurrent disease. (G) In 76 breast cancer patients who received first-line CMF-based chemotherapy for recurrent disease. (H) In 150 breast cancer patients who received first-line anthracycline based chemotherapy for recurrent disease. Differences between the survival curves were calculated with the 3-sample logrank test.
Table 2. Univariate Cox regression analysis to evaluate the association of calculated APOBEC3B copy number values with the length of MFS.
| Study cohort | N Patients | N Events | Univariate analysis | |
|---|---|---|---|---|
| HR (95% CI) | P-value | |||
| LNN+LNP | ||||
| All patients | 1,604 | 686 | 0.95 (0.88–1.02) | 0.17 |
| LNN | ||||
| All patients | 1,076 | 366 | 1.00 (0.90–1.11) | 0.96 |
| ER+ patients | 769 | 263 | 0.93 (0.82–1.06) | 0.29 |
| ER- patients | 300 | 101 | 1.13 (0.94–1.35) | 0.20 |
N, number of; HR, hazard ratio; CI, confidence interval; LNN, lymph node negative; LNP, lymph node positive.
Note: ER status was not available for 7 out of 1,076 patients.
To evaluate the predictive value of APOBEC3B copy numbers, we had two cohorts of breast cancer patients available that were treated with first-line therapy for recurrent disease. First, we evaluated whether the calculated APOBEC3B copy number values could predict the clinical outcome of first-line tamoxifen therapy in a cohort of 329 hormone-naive breast cancer patients with ER-positive primary breast cancer. No significant association was observed between APOBEC3B copy number and the ORR for tamoxifen therapy (OR = 0.88, 95% CI = 0.69–1.13, P = 0.31; Table 3). Moreover, APOBEC3B copy number status was not associated with the length of PFS (P = 0.25; Fig 2E), nor were calculated APOBEC3B copy number values associated with the length of PFS (HR = 1.00, 95% CI = 0.88–1.14, P = 0.96; Table 4) in this cohort. Thus, APOBEC3B copy number is not a suitable biomarker to predict the type of response to tamoxifen therapy.
Table 3. Univariate logistic regression analysis of the overall response rate in patients treated with first-line tamoxifen and in patients treated with first-line chemotherapy for recurrent disease.
| Study cohort | N Patients | Univariate analysis | |
|---|---|---|---|
| OR (95% CI) | P-value | ||
| First-line tamoxifen | |||
| All patients | 329 | 0.88 (0.69–1.13) | 0.31 |
| First-line chemotherapy | |||
| All patients | 224 | 0.96 (0.71–1.31) | 0.80 |
| CMF-treated patients | 75 | 1.25 (0.70–2.21) | 0.45 |
| Anthracyclin-treated patients | 149 | 0.79 (0.53–1.16) | 0.22 |
N, number of; OR, odds ratio; CI, confidence interval; CMF, cyclophosphamide/methotrexate/5-fluorouracil.
Note: the type of response was ambiguous for 2 patients
Table 4. Univariate Cox regression analysis to evaluate the association of calculated APOBEC3B copy number values with the length of PFS.
| Study cohort | N Patients | N Events | Univariate analysis | |
|---|---|---|---|---|
| HR (95% CI) | P-value | |||
| First-line tamoxifen | ||||
| All patients | 329 | 304 | 1.00 (0.88–1.14) | 0.96 |
| First-line chemotherapy | ||||
| All patients | 226 | 138 | 1.06 (0.88–1.29) | 0.53 |
| CMF-treated patients | 76 | 52 | 0.91 (0.65–1.28) | 0.58 |
| Anthracyclin-treated patients | 150 | 86 | 1.19 (0.93–1.52) | 0.17 |
N, number of; HR, hazard ratio; CI, confidence interval; CMF, cyclophosphamide/methotrexate/5-fluorouracil.
Note: The length of PFS was censored at two months after the start of consolidation therapy.
Next, we evaluated whether calculated APOBEC3B copy number values could predict the outcome of first-line chemotherapy in a cohort of 226 breast cancer patients. The calculated APOBEC3B copy number values were, however, not found to be associated with the ORR for chemotherapy (OR = 0.96, 95% CI = 0.71–1.31, P = 0.80; Table 3). In addition, neither APOBEC3B copy number status, nor calculated APOBEC3B copy number values were associated with the length of PFS in these patients (P = 0.42; Fig 2F and HR = 1.06, 95% CI = 0.88–1.29, P = 0.53; Table 4, respectively). The lack of a significant association with the ORR to chemotherapy and the length of PFS was also observed when performing subgroup analysis by type of chemotherapy (i.e. CMF versus anthracyclines; Table 3, Fig 2G and 2H, Table 4). APOBEC3B copy number is thus also not a predictive biomarker for the type of response to chemotherapy.
In a previous study, we had analyzed APOBEC3B mRNA expression in 1,491 breast cancer patients and found that high APOBEC3B expression had prognostic value and was associated with poor outcome in untreated LNN patients with ER-positive breast cancer [15]. However, we did not find any association between APOBEC3B copy numbers and clinical outcome in the current study. As both APOBEC3B expression and APOBEC3B copy number have been associated with a hypermutator phenotype [13,14,24], we attempted to clarify this discrepancy by evaluating the correlation between APOBEC3B copy number and APOBEC3B mRNA expression. Out of the 1,756 patients for whom we performed APOBEC3B copy number analysis in the current study, we had APOBEC3B mRNA expression data available for 1,132 patients. Interestingly, although a correlation among APOBEC3B copy number and mRNA expression was observed, the correlation coefficient was low (spearman’s rho = 0.26, P = 2.2*10−16). This suggests that other mechanisms exist that affect APOBEC3B mRNA expression besides APOBEC3B copy number.
Discussion
APOBEC3B mRNA expression is upregulated in multiple tumor types and this has been shown to correlate with an increased mutational load, particularly an increase in C>T transversions [13,14]. In line with these findings, increased expression of APOBEC3B mRNA was associated with a poor prognosis in ER-positive breast cancer [15]. At the same time, the 29.5 kb deletion polymorphism of APOBEC3B was found to be associated with the increased breast cancer risk in different populations [19–21], although these findings were not confirmed in a Swedish study [22]. Similar to APOBEC3B overexpression, the APOBEC3B deletion has been shown to correlate with an increased mutational load [24]. These findings may seem paradoxical, as loss of APOBEC3B should decrease the mutational load. However, the APOBEC3B deletion polymorphism not just deletes APOBEC3B. It also generates a novel fusion transcript (i.e. APOBEC3A under the control of the 3’UTR of APOBEC3B) [18]. Consequently, APOBEC3A mRNA was shown to become more stable, resulting in higher levels of APOBEC3A and, since APOBEC3A is a more efficient hypermutator than APOBEC3B, more severe DNA damage [23]. Thus, although the molecular mechanisms behind overexpression of APOBEC3B and the APOBEC3B deletion polymorphism are very different, they both result in a hypermutator phenotype. The hypothesis that the APOBEC3B deletion polymorphism thus may also be associated with clinical outcome is therefore plausible.
In a study by Gohler et al., the APOBEC3B deletion polymorphism was however, not associated with breast cancer specific survival in 782 breast cancer cases [22]. Moreover, Cescon et al. showed that the deletion was not associated with recurrence after treatment for early breast cancer in METABRIC [16]. Because both studies also included patients that were treated, no distinction could be made between pure prognosis and therapy response. In the current study, we examined separately the prognostic and predictive value of APOBEC3B copy number in a total of 1,756 breast cancer patients. No association between APOBEC3B copy numbers and the length of MFS was found among 1,076 LNN patients who had not received adjuvant systemic treatment (Fig 2B, Table 2). In addition, an association with the length of MFS was also not observed among ER-positive or ER-negative breast cancer patients. These results imply that APOBEC3B copy number is not a prognostic biomarker for breast cancer. The association between APOBEC3B copy number and the response to treatment in breast cancer patients that received either first-line tamoxifen or chemotherapy for recurrent disease was also evaluated. However, we found no association between the type of response for either tamoxifen or chemotherapy and APOBEC3B copy number. Thus, besides not having any prognostic value, APOBEC3B copy numbers also do not appear to have a predictive value for breast cancer patients.
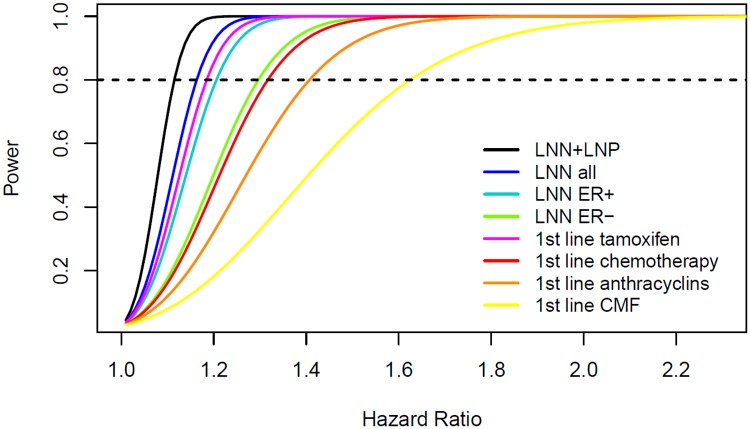
The copy number analyses in the adjuvant setting, except for the ER-negative LNN analysis, had sufficient power to conclude that there is no or only a marginal prognostic effect of APOBEC3B copy numbers in breast cancer patients (Fig 3). For the copy number analysis in the advanced setting involving patients treated with first-line tamoxifen, we could draw the same conclusion (Fig 3). However, for the copy number analysis involving patients treated with first-line chemotherapy, especially in the subgroup analyses by type of chemotherapy, we cannot exclude a modest effect of APOBEC3B copy numbers on the length of PFS (Fig 3). Therefore, replication of the results we observed in the advanced chemotherapy setting in a larger sample size or population with a higher prevalence of the APOBEC3B deletion polymorphism would be needed.
Fig 3. Power as a function of the hazard ratio for APOBEC3B copy number in each of the analyzed subgroups.
The dashed horizontal line crosses the curve of each subgroup at the minimal hazard ratio for which we had 80% power in our APOBEC3B copy number analyses.
Another note is that the assay we used to determine APOBEC3B copy numbers does not discriminate between germline and tumor-specific (i.e. somatic) APOBEC3B deletion. This has no consequence for the accuracy of the germline copy number determination except for patients who carry a one-copy deletion and have strong amplification of the remaining APOBEC3B locus. Patients who do not carry the germline deletion and have either a tumor-specific homozygous deletion or a heterozygous deletion of APOBEC3B in combination with a high tumor cell percentage may also be misclassified. In this respect, the observed amplification of the APOBEC3B allele is rather a somatic event than the result of an alteration in the germline. To date, there has been no report of the APOBEC3B locus being amplified in the germline. Interestingly, in a publically available SNP array data set of 344 breast cancers (accession number EGAS00001001178 [33]), APOBEC3A copy number status was equal to APOBEC3B copy number status in all tumors, suggesting that APOBEC3A is always amplified or deleted simultaneously with APOBEC3B during breast tumorigenesis.
In our Kaplan-Meier survival analysis we grouped patients with one-copy deletion and two-copy deletions together to provide a visualization of the estimated survival curves of patients with a deleted, balanced or amplified APOBEC3B gene. Unfortunately, the population frequency of the 29.5 kb deletion polymorphism of APOBEC3B was too low in this Dutch cohort to analyze patients who carry a two-copy deletion separately. To illustrate this with a power calculation: we would need a sample size of 4,024 or 11,759 LNN patients to ensure a minimally detectable hazard ratio of 2 or 1.5, respectively, using a power of 80% and an alpha of 0.05. For this reason, evaluation of the clinical value of the two-copy deletion should preferably be done in breast cancer patients from the Asian or American population (i.e. population frequency of 36.9% and 57.7%, respectively)[18].
The observation that the APOBEC3B deletion polymorphism does not seem to have any impact on the clinical outcome for breast cancer patients, whereas increased expression of APOBEC3B mRNA does, strengthens the evidence that there are two different molecular mechanisms in place. Moreover, elevated levels of APOBEC3B mRNA have been shown to associate with cellular proliferation, whereas the APOBEC3B deletion polymorphism associated with activation of immune-related genes [16]. Interestingly, increased lymphocytic infiltration has been associated with a favorable outcome in some, but not all subtypes of breast cancer [34,35]. This is in contrast to what has been observed for increased APOBEC3B mRNA expression, which associated with increased proliferation and poor outcome [15].
Although we observed a correlation between APOBEC3B copy number and mRNA expression, this correlation was rather weak. As a consequence, the prognostic effect of increased levels of APOBEC3B mRNA is not necessarily caused by increased APOBEC3B copy numbers. Other mechanisms thus exist that elevate APOBEC3B mRNA levels. Recently, it was shown that APOBEC3B interacts with ER to bind to ER binding sites, where it generates C to U transitions. Furthermore, the presence of APOBEC3B was necessary for histone modification, but also in order to recruit chromatin remodeling factors to ER binding sites [36]. This may very well explain why elevated levels of APOBEC3B mRNA are only prognostic in ER-positive breast cancer patients. Although the APOBEC3B deletion polymorphism does not have any prognostic or predictive value, it does appear to contribute to breast tumorigenesis by conferring an increased risk to develop breast cancer. However, still little is known regarding the role of APOBEC3A in breast cancer. Therefore, more studies should be done in order to investigate the precise effect of the APOBEC3B deletion polymorphism and APOBEC3A-B hybrid transcript resulting from this deletion. This will provide more insight into APOBEC-mediated hypermutation.
Supporting Information
(PDF)
(XLSX)
Acknowledgments
We thank Marcel Smid for assistance with bioinformatics analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by Cancer Genomics Netherlands (CGC.nl) and a grant for the Netherlands Organization of Scientific Research (NWO). JL received a scholarship from the China Scholarship Council (Beijing, China). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat A, Pineda E, Adamo B, Galvan P, Fernandez A, Gaba L, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015; [DOI] [PubMed] [Google Scholar]
- 5.Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68(9):3108–3114. 10.1158/0008-5472.CAN-07-5644 [DOI] [PubMed] [Google Scholar]
- 6.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993. 10.1016/j.cell.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2(e00534 10.7554/eLife.00534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22(2):367–377. [DOI] [PubMed] [Google Scholar]
- 10.Harris RS Molecular mechanism and clinical impact of APOBEC3B-catalyzed mutagenesis in breast cancer. Breast Cancer Res. 2015;17(8 10.1186/s13058-014-0498-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103(23):8780–8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281(31):22161–22172. [DOI] [PubMed] [Google Scholar]
- 13.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494(7437):366–370. 10.1038/nature11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns MB, Temiz NA, Harris RS Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45(9):977–983. 10.1038/ng.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sieuwerts AM, Willis S, Burns MB, Look MP, Meijer-Van Gelder ME, Schlicker A, et al. Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm Cancer. 2014;5(6):405–413. 10.1007/s12672-014-0196-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cescon DW, Haibe-Kains B, Mak TW APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc Natl Acad Sci U S A. 2015;112(9):2841–2846. 10.1073/pnas.1424869112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuboi M, Yamane A, Horiguchi J, Yokobori T, Kawabata-Iwakawa R, Yoshiyama S, et al. APOBEC3B high expression status is associated with aggressive phenotype in Japanese breast cancers. Breast Cancer. 2015; [DOI] [PubMed] [Google Scholar]
- 18.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3(4):e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long J, Delahanty RJ, Li G, Gao YT, Lu W, Cai Q, et al. A common deletion in the APOBEC3 genes and breast cancer risk. J Natl Cancer Inst. 2013;105(8):573–579. 10.1093/jnci/djt018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xuan D, Li G, Cai Q, Deming-Halverson S, Shrubsole MJ, Shu XO, et al. APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis. 2013;34(10):2240–2243. 10.1093/carcin/bgt185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rezaei M, Hashemi M, Hashemi SM, Mashhadi MA, Taheri M APOBEC3 Deletion is Associated with Breast Cancer Risk in a Sample of Southeast Iranian Population. Int J Mol Cell Med. 2015;4(2):103–108. [PMC free article] [PubMed] [Google Scholar]
- 22.Gohler S, Da Silva Filho MI, Johansson R, Enquist-Olsson K, Henriksson R, Hemminki K, et al. Impact of functional germline variants and a deletion polymorphism in APOBEC3A and APOBEC3B on breast cancer risk and survival in a Swedish study population. J Cancer Res Clin Oncol. 2015; [DOI] [PubMed] [Google Scholar]
- 23.Caval V, Suspene R, Shapira M, Vartanian JP, Wain-Hobson S A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3'UTR enhances chromosomal DNA damage. Nat Commun. 2014;5(5129 10.1038/ncomms6129 [DOI] [PubMed] [Google Scholar]
- 24.Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet. 2014;46(5):487–491. 10.1038/ng.2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foekens JA, Portengen H, Look MP, van Putten WL, Thirion B, Bontenbal M, et al. Relationship of PS2 with response to tamoxifen therapy in patients with recurrent breast cancer. Br J Cancer. 1994;70(6):1217–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foekens JA, Portengen H, van Putten WL, Peters HA, Krijnen HL, Alexieva-Figusch J, et al. Prognostic value of estrogen and progesterone receptors measured by enzyme immunoassays in human breast tumor cytosols. Cancer Res. 1989;49(21):5823–5828. [PubMed] [Google Scholar]
- 27.van Agthoven T, Sieuwerts AM, Meijer-van Gelder ME, Look MP, Smid M, Veldscholte J, et al. Relevance of breast cancer antiestrogen resistance genes in human breast cancer progression and tamoxifen resistance. J Clin Oncol. 2009;27(4):542–549. 10.1200/JCO.2008.17.1462 [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. [DOI] [PubMed] [Google Scholar]
- 29.Hayward JL, Carbone PP, Heuson JC, Kumaoka S, Segaloff A, Rubens RD Assessment of response to therapy in advanced breast cancer: a project of the Programme on Clinical Oncology of the International Union Against Cancer, Geneva, Switzerland. Cancer. 1977;39(3):1289–1294. [DOI] [PubMed] [Google Scholar]
- 30.European Organization for Research and Treatment of Cancer, Breast Cancer Cooperative Group Manual for clinical research and treatment in breast cancer. Excerpta Medica Almere, the Netherlands. 2000;116–117.
- 31.McShane LM, Altman DG, S W., Taube SE, Gion M, Clark GM, et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;97(16):1180–1184. [DOI] [PubMed] [Google Scholar]
- 32.Sieuwerts AM, Meijer-van Gelder ME, Timmermans M, Trapman AM, Garcia RR, Arnold M, et al. How ADAM-9 and ADAM-11 differentially from estrogen receptor predict response to tamoxifen treatment in patients with recurrent breast cancer: a retrospective study. Clin Cancer Res. 2005;11(20):7311–7321. [DOI] [PubMed] [Google Scholar]
- 33.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. 10.1038/nature17676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02–98. J Clin Oncol. 2013;31(7):860–867. 10.1200/JCO.2011.41.0902 [DOI] [PubMed] [Google Scholar]
- 35.Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol. 2014;25(8):1544–1550. 10.1093/annonc/mdu112 [DOI] [PubMed] [Google Scholar]
- 36.Periyasamy M, Patel H, Lai CF, Nguyen VT, Nevedomskaya E, Harrod A, et al. APOBEC3B-Mediated Cytidine Deamination Is Required for Estrogen Receptor Action in Breast Cancer. Cell Rep. 2015;13(1):108–121. 10.1016/j.celrep.2015.08.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.