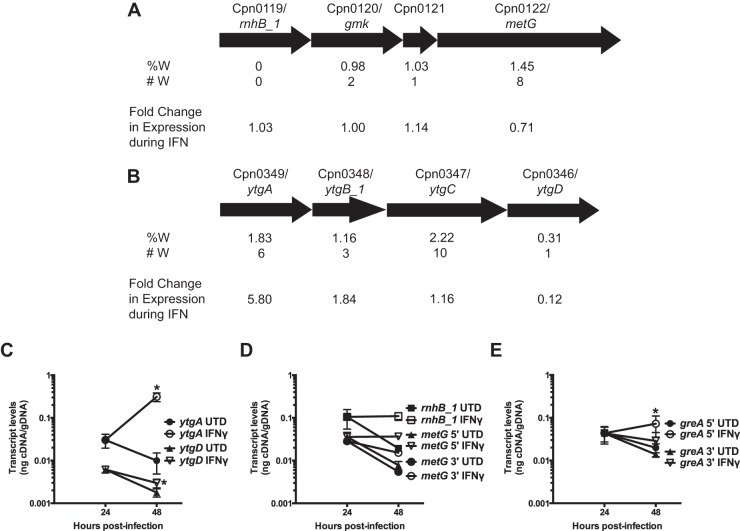
FIG 5.
Tryptophan codons have destabilizing effects on downstream transcripts. Shown is determination of the effect of Trp (W) codons on transcript levels within operons and large genes in C. pneumoniae during IFN-γ-mediated persistence. (A and B) Schematic representation of the (A) ytg and (B) rnhB_1 operons, including the percentage (%W) and number (#W) of Trp codons within the open reading frames. Arrow sizes represent the relative sizes of the genes in the operons. Also shown is the fold change in transcript levels during IFN-γ persistence between 24 and 48 h postinfection, as measured by microarray (42). (C to E) Nucleic acid samples were collected from untreated (solid symbols) and IFN-γ-treated (open symbols) C. pneumoniae-infected HEp2 cells at 24 and 48 h postinfection. Transcript levels for the indicated genes were measured by RT-qPCR and normalized to genomic DNA (nanograms of cDNA/gDNA). (C) Quantification of transcript levels of ytgA and ytgD. (D) Quantification of transcript levels of rnhB_1 and metG. Two different primer sets were used to quantify metG transcripts: one targeting the 5′ end of the gene and another the 3′ end. (E) Quantification of transcript levels of greA using primer sets designed against the 5′ and 3′ ends of the gene. (See the text for more detail.) All transcript data are the average from 3 experiments (except metG 3′, where n = 2) with standard deviations displayed. *, P ≤ 0.05 for 48-hpi IFN-γ versus 24-hpi IFN-γ.