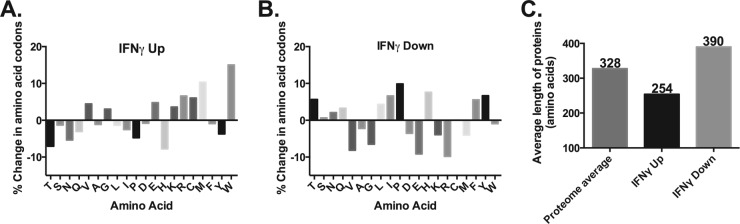
FIG 6.
Codon content analysis of C. pneumoniae transcript data during IFN-γ-mediated persistence reveals an enrichment of tryptophan codon-containing genes in the upregulated data set. Microarray data from IFN-γ-treated C. pneumoniae were analyzed based on the codon content of the genes that were at least 2-fold increased (A [IFNγ Up]) or 2-fold decreased (B [IFNγ Down]). The percentage change in the codon content is reflected on the y axis, whereas the individual single-letter code for the amino acid is listed on the x axis. The 0 value reflects the average C. pneumoniae codon content of the 24-h postinfection (i.e., RB profile) transcript levels. (C) For the data in panels A and B, the average length of the proteins encoded by the differentially transcribed genes was plotted and compared to the proteome average (328 amino acids).