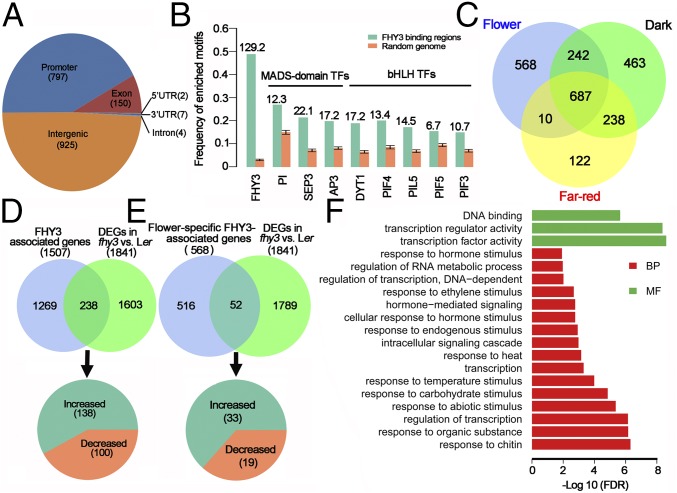
Fig. 2.
Genome-wide identification of FHY3 binding sites and target genes. (A) Classification of FHY3 binding sites in the Arabidopsis genome. The numbers of binding sites are indicated in parentheses. (B) The binding motifs of several TFs were significantly enriched around the FHY3 binding peaks compared with randomly selected genomic regions. The numbers on the top of columns are z-scores computed from the permutation test. A z-score of 2 or above is considered statistically significant. (C) Venn diagram showing the number and overlap of FHY3-associated genes in flower and seedling under D and FR conditions. (D and E) The FHY3 ChIP-seq data and RNA-seq data were compared to identify FHY3 target genes (D) and flower-specific FHY3 target genes (E). (F) Enrichment of GO terms among flower-specific FHY3 target genes. BP, biological process; MF, molecular function.