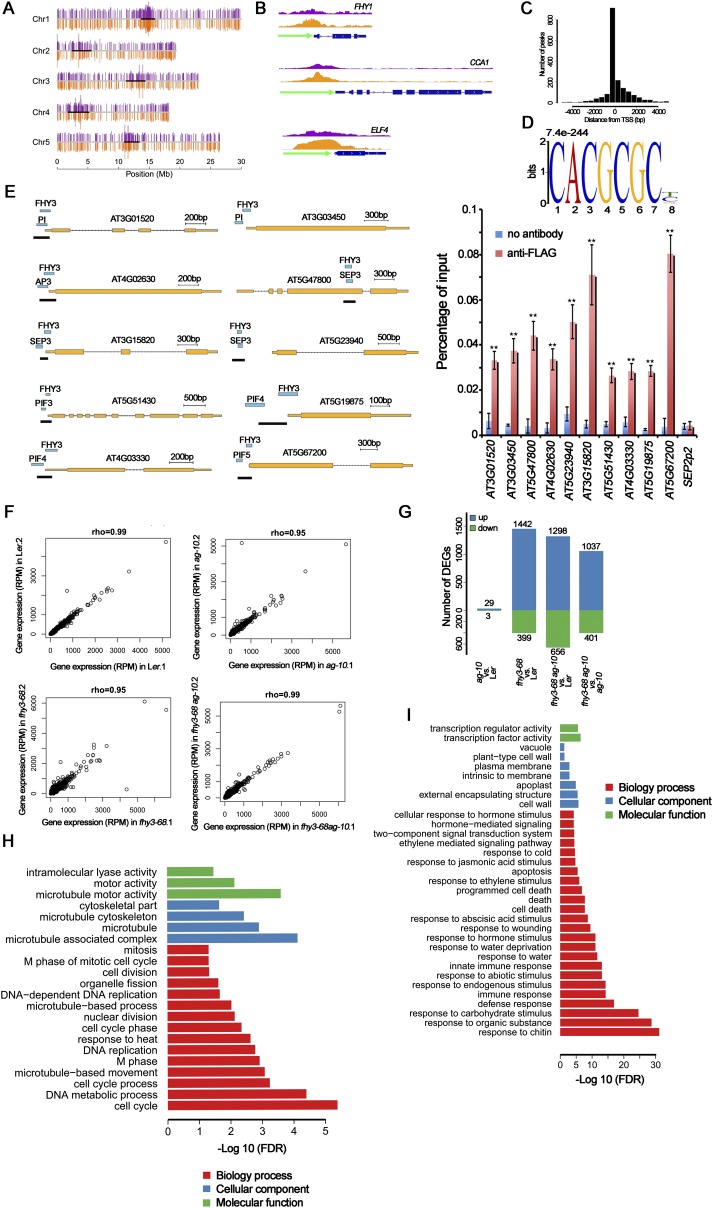
Fig. S3.
ChIP-seq and RNA-seq analysis of FHY3 binding sites. (A) Distribution of FHY3 binding sites on the five Arabidopsis chromosomes. The top purple bars and bottom orange bars on each chromosome represent the positions of the FHY3 binding sites from two biological replicates. The scale at the bottom indicates chromosome positions. (B) FHY3 binding sites were found in the promoter region of three known FHY3 target genes FHY1, CCA1, and ELF4. FLAG-FHY3 peaks (purple and orange) from two biological replicates and gene structure are shown in the top, middle, and bottom rows, respectively. (C) FHY3 binding sites are highly enriched in the regions around the TSS. (D) The typical FBS motif (CACGCGC, E-value = 7.4e-244) was identified as a statistically significant motif in the FHY3 binding regions in flower. (E) ChIP-PCR to verify the colocalization of FHY3 and indicated TFs. FLAG-FHY3 and TFs peaks, gene structures and the regions examined by ChIP (marked by black lines) are shown (Left). ChIP to measure FHY3 occupancy at loci in 35S:3FLAG-FHY3-3HA fhy3-4 inflorescences (Right). SEP2P2 served as a negative control. Error bars represent SD from three biological replicates. **P < 0.01 compared with no antibody (negative control). (F) Scatterplots of gene expression data from two replicates for each sample. Expression level was normalized to reads per million (RPM). Spearman correlation coefficients were calculated as an indicator of reproducibility between replicates. (G) Number of DEGs identified in pairwise comparisons of RNA-seq data. (H and I) Significantly enriched GO terms in the down-regulated genes (H) and up-regulated genes (I) in fhy3-68 vs. Ler (genes from G).