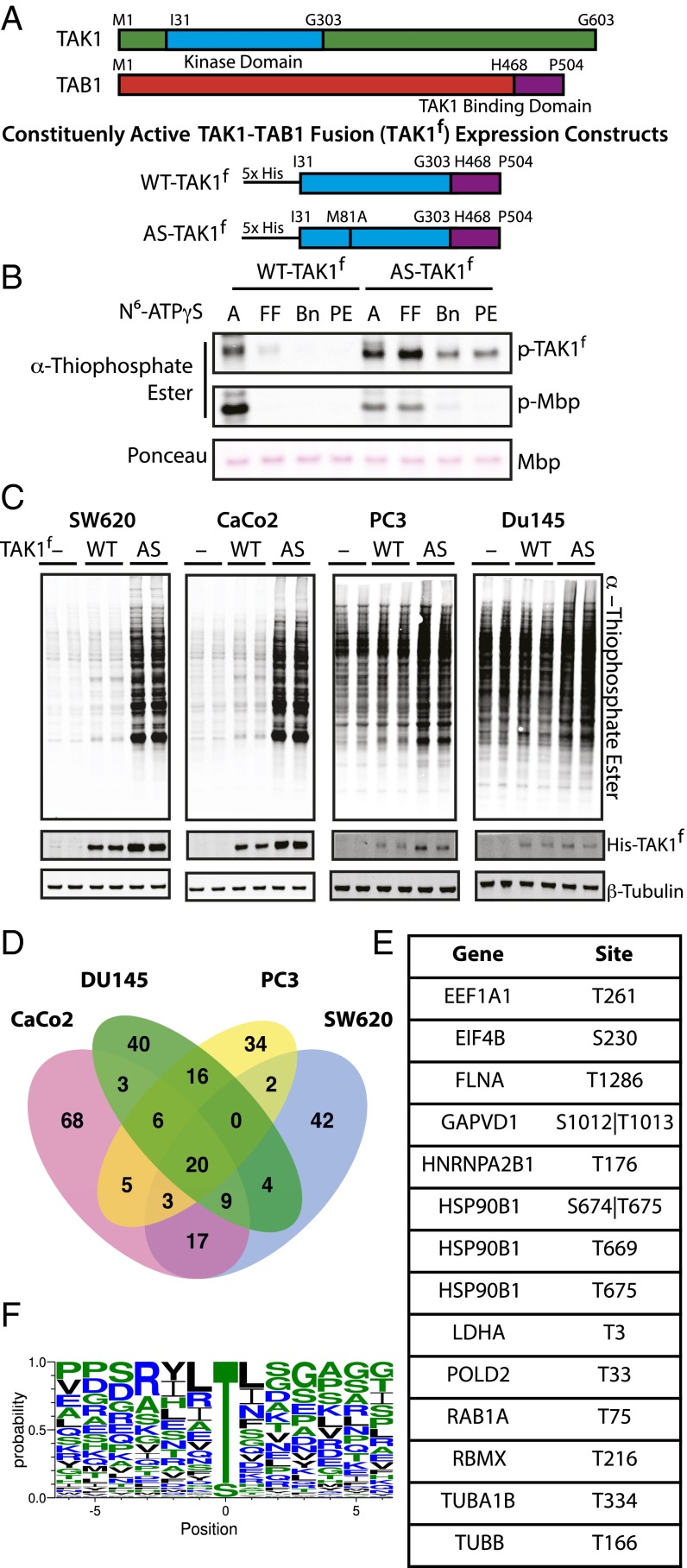
Fig. 1.
Characterization of AS-TAK1f and identification of TAK1 substrates. (A) Schematic of full-length TAK1, TAB1, and constituently active fusion constructs of WT-TAK1f and AS-TAK1f used for protein purification. (B) In vitro kinase assay with TAK1f, MBP, and bulky N6-substituted ATPγS analogs (A, ATPγS; Bn, N6 benzyl; FF, N6 furfuryl; PE, N6 phenethyl). Thiophosporylation was evaluated by Western blot. (C) Lysate from SW620, CaCo2, PC3, and Du145 cells were labeled with no kinase (−), His-tagged WT-TAK1f (WT), or His-tagged AS-TAK1f (AS) in biological duplicate. (D) Venn diagrams of phosphoproteins identified in four cell cancer lines, colorectal (SW620, CaCo2) and pancreatic (DU145, PC3). (E) Phosphosites exclusive to and identified in all eight individual AS-TAK1f–labeled samples. (F) TAK1 consensus motif derived from all phosphopeptides identified in ≥2 samples.