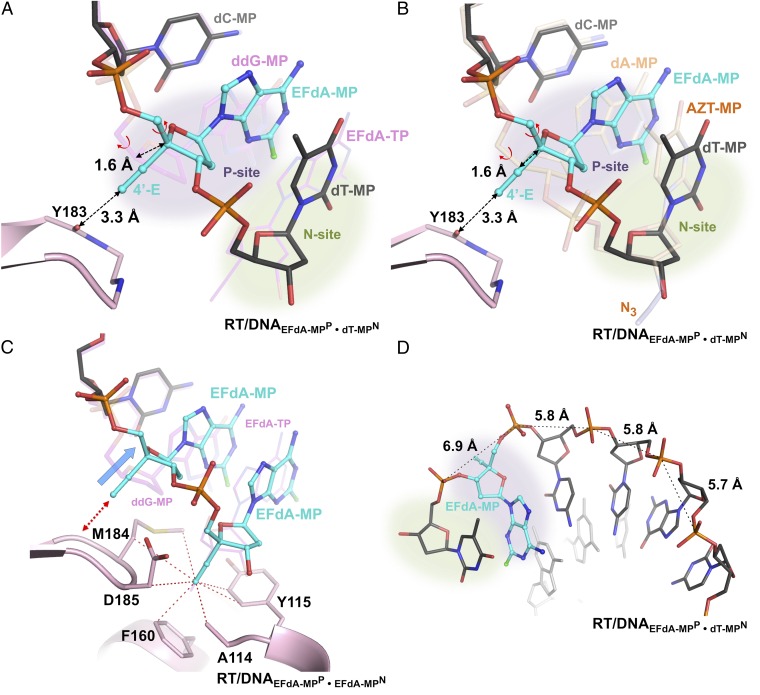
Fig. 5.
Structural changes in EFdA-containing complexes. (A) Superposition of N-complex RT/DNAEFdA-MPP•dT-MPN and ternary complex RT/DNAddG-MPP/EFdA-TP (magenta DNA). (B) Superposition of two N complexes of RT, in the presence or absence of EFdA-MP at the P site [RT/DNAEFdA-MPP•dT-MPN and RT/DNAdA-MPP•AZT-MPN (PDB ID 1N6Q in light brown) (17)]. In both A and B, without the localized 1.6-Å primer shift (marked dotted arrow) of EFdA-MPP, the currently observed 3.3-Å distance between the 4′-E and main-chain carbonyl of Y183 would have been highly unfavorable at ∼1.7 Å. This shift is accomplished through a change in the γ-torsion angle about the C4′-C5′ (Fig. S6A vs. Fig. S6B; Table S1). (C) Primer distortion due to interactions of P-site EFdA-MP of RT/DNAEFdA-MPP•EFdA-MPN with Y183 (red arrow) compared with RT/DNAddG-MPP/EFdA-TP (magenta DNA); hydrophobic interactions of the 4′-E at the active-site hydrophobic pocket (red dotted lines connect atoms within 4 Å). (D) The change in γ results in increased distance between neighboring phosphates near the polymerase active site.