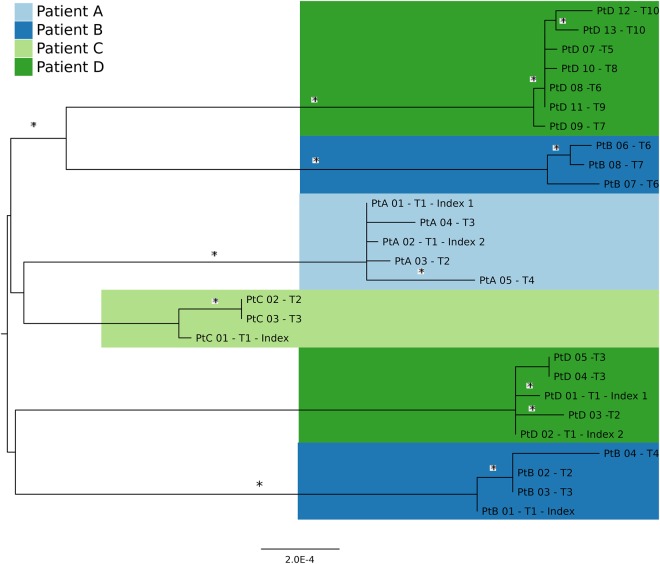
Figure 1.
Maximum likelihood phylogenetic tree inferred from single-nucleotide polymorphism (SNP) alignment of 27 methicillin-resistant Staphylococcus aureus USA300 isolates and colored by patient. Phylogeny was inferred using RAxML v8.0.0, using the ASC_GTRGAMMA nucleotide substitution model with ascertainment bias correction and 100 bootstrap replicates. The tree is midpoint rooted, and clades with bootstrap values of >80% are denoted with an asterisk. The phylogeny illustrates that patients B and D had strain replacement events in which a separate USA300 population replaced their incident USA300 population. Separate strain acquisitions are inferred from the temporality of isolate collection and phylogenetic data illustrating distinct, well-supported monophyletic clades for each USA300 population.