Abstract
Diazo groups are found in a range of natural products that possess potent biological activities. Despite longstanding interest in these metabolites, diazo group biosynthesis is not well understood, in part because of difficulties in identifying specific genes linked to diazo formation. Here we describe the discovery of the gene cluster that produces the o-diazoquinone natural product cremeomycin and its heterologous expression in Streptomyces lividans. We have used stable isotope feeding experiments and in vitro characterization of biosynthetic enzymes to decipher the order of events in this pathway and establish that diazo construction involves late-stage N–N bond formation. This work represents the first successful production of a diazo-containing metabolite in a heterologous host, experimentally linking a set of genes with diazo formation.
Keywords: natural products, biosynthesis, diazo compounds, enzymes, heterologous expression
Table of Contents
An experimental link to Nature’s reactivity: The heterologous production of the natural product cremeomycin experimentally links genes with diazo biosynthesis for the first time. Characterization of the order of events in the pathway reveals the timing of diazo assembly. Comparison with other biosynthetic gene clusters suggests bacteria install this reactive group in multiple ways.
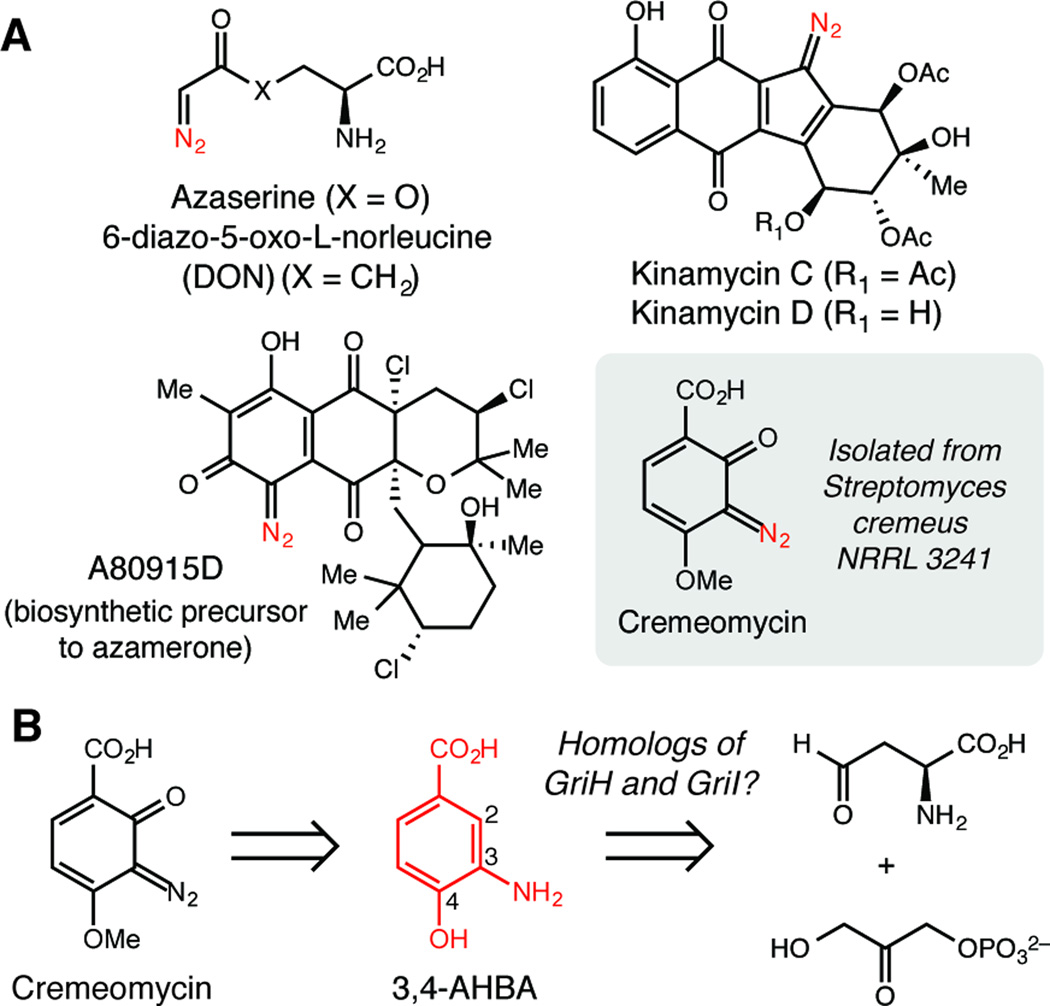
Diazo-containing compounds are exceptionally important reagents with unique reactivity that facilitates 1,3-dipolar cycloadditions, carbene insertions, and alkylations. Such broad reactivity is useful for applications in both synthetic chemistry and chemical biology.[1] This functional group is also found in diverse natural products that have interesting biological activity (Figure 1A).[2] The first diazo-containing metabolites discovered were the modified α-amino acids azaserine, 6-diazo-5-oxo-L-norleucine (DON), and azotomycin, which were isolated from several Streptomyces strains in the 1950s.[3] These L-glutamine mimics gained considerable attention for their therapeutic potential and were evaluated clinically.[4] More recently, studies of the diazofluorene antitumor antibiotics kinamycin C and lomaiviticin A showed that both diazo groups are required for the remarkably enhanced cytotoxicity and DNA-cleaving activity of the latter molecule.[5]
Figure 1.
Cremeomycin is an o-diazoquinone natural product. A) Selected diazo-containing secondary metabolites. B) Biosynthetic hypothesis for cremeomycin assembly.
Despite sustained interest in diazo-containing natural products from both organic and biological chemists, neither the genes nor the enzymatic transformations involved in diazo assembly have been characterized experimentally. Understanding the genetic and biochemical basis for diazo installation would fill a void in our understanding of enzymatic bond constructions and could facilitate biocatalytic and synthetic biology efforts to both yield reagents for synthetic chemistry and enable the synthesis of clinically relevant molecules.
Insights into diazo biosynthesis have been gained from feeding studies with the metabolites kinamycin D and A80915D, a biosynthetic precursor to the pyridazine-containing metabolite azamerone. Supplementation of culture media with 15N-labeled inorganic nitrogen sources (A80915D) and a 2H-labeled putative intermediate (kinamycin D) has established that diazo installation proceeds via N–N bond formation between two nitrogen-containing precursors, not through dinitrogen fixation.[6] Recently, our group and others used bioinformatics to identify a set of genes in the kinamycin and lomaiviticin gene clusters that we propose are responsible for diazo formation.[7] Though the contexts of these genes strongly suggest a role in diazo assembly, their functions have not yet been validated experimentally in vivo or in vitro. In particular, the inability to reconstitute the biosynthetic pathways of these molecules and other diazo containing natural products in heterologous hosts has hindered efforts to link specific genes to the construction of this functional group.[6a]
Here we report the discovery and heterologous expression of the biosynthetic gene cluster responsible for producing the o-diazoquinone natural product cremeomycin. This work represents the first successful heterologous production of a diazo-containing metabolite, thus establishing an experimental link between a set of genes and diazo biosynthesis for the first time. Using in vitro enzyme characterization and stable isotope feeding studies, we establish the order of events in cremeomycin biosynthesis and show that diazo installation requires late-stage N–N bond formation. The success of our efforts represents a significant advance in our understanding of the genetic basis for diazo assembly and highlights this pathway as an excellent system for further mechanistic investigations.
Cremeomycin is a light-sensitive o-diazoquinone with antibacterial and antiproliferative effects.[8] It was isolated by the Upjohn Company in 1967 from the soil actinomycete Streptomyces cremeus NRRL 3241.[8a] A total synthesis of cremeomycin has also been reported.[9] We used a chemically guided genome mining approach to identify the cremeomycin (cre) biosynthetic gene cluster based on the previously-proposed hypothesis that 3-amino-4-hydroxybenzoic acid (3,4-AHBA) is a biosynthetic intermediate (Figure 1B).[8b] This aromatic metabolite is used in multiple natural product biosynthetic pathways (Figure S12). In grixazone biosynthesis, 3,4-AHBA is synthesized from the primary metabolites dihydroxyacetone phosphate and 4-aspartate semialdehyde by the enzymes GriI and GriH.[10] GriI is homologous to class I aldolases, and it catalyzes C–C bond formation to generate an intermediate that is cyclized by GriH to afford 3,4-AHBA.
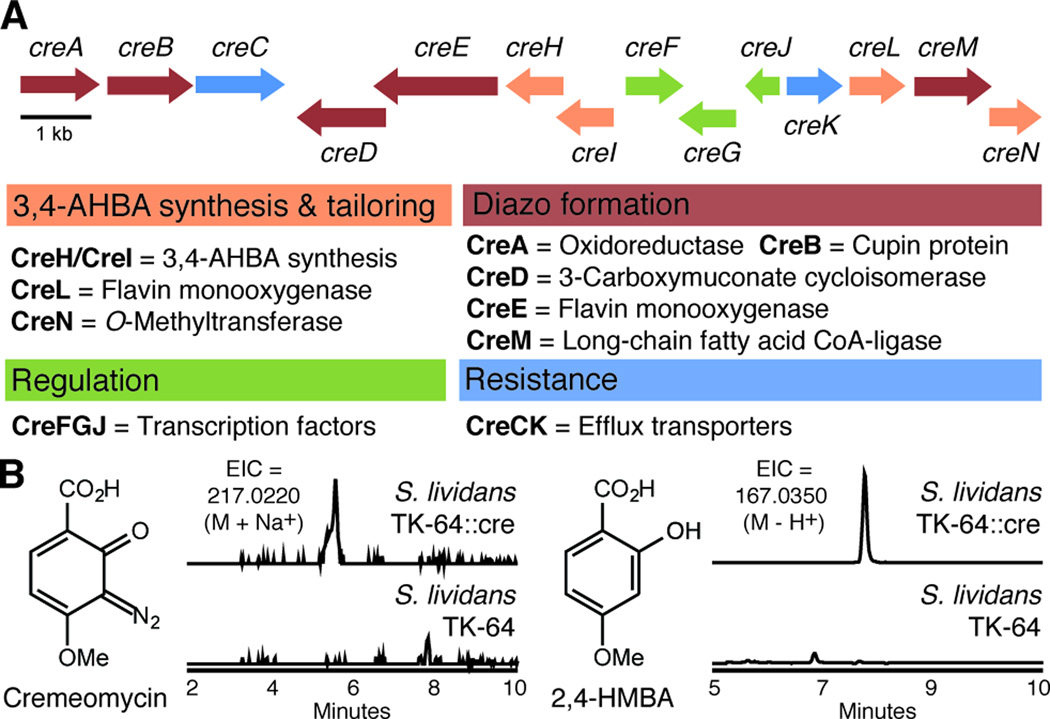
To determine whether S. cremeus possessed these 3,4-AHBA-synthesizing enzymes, we used degenerate PCR to amplify putative 3,4-AHBA synthase (griH) homologs from genomic DNA.[11] A PCR product with high identity to griH was obtained, suggesting that S. cremeus could synthesize 3,4-AHBA. With initial support for our biosynthetic hypothesis, we sequenced the genome of S. cremeus. BLAST searches using the griH fragment revealed an open reading frame (ORF) encoding a griH homolog (creH) on a 370 kilobase (kb) scaffold. In close proximity to this gene were three additional ORFs encoding enzymes with predicted functions consistent with a role in cremeomycin assembly: an aldolase homolog (creI), a Class A flavin-dependent aromatic monooxygenase (creL), and an S-adenosylmethionine (SAM)-dependent O-methyltransferase (creN). To determine the cluster boundaries we searched the adjacent regions of the scaffold for synteny with the genomes of Streptomyces not known to produce cremeomycin. This analysis revealed high conservation on either side of an 18 kb region encoding 14 ORFs (creA-creN), which we termed the cre gene cluster (Figure 2A & Table S1). In addition to the genes predicted to be responsible for 3,4-AHBA synthesis and tailoring (creHILN), there were five additional genes encoding enzymes that could be involved in diazo formation (creABDEM). The predicted functions of some of the encoded enzymes (N-hydroxylation, activation with ATP) are consistent with the chemistry required for constructing a diazo group from two separate nitrogen-containing metabolites.
Figure 2.
Identification and heterologous expression of the cremeomycin (cre) biosynthetic gene cluster. A) The cre biosynthetic gene cluster. Refer to SI for complete annotation. B) Heterologous expression of the cre cluster in S. lividans TK-64 affords production of cremeomycin and 2,4-HMBA. Extracted ion chromatograms (EIC) are shown.
We confirmed the link between the cre gene cluster and cremeomycin production through heterologous expression in Streptomyces lividans TK-64. The integrative plasmid pCre was constructed from a genomic library of S. cremeus and introduced into S. lividans TK-64 via standard protoplast transformation. Culturing of this strain in the absence of light in ISP1 media and analysis of organic extracts of the culture broth using liquid chromatography-mass spectrometry (LC-MS) and LC-MS/MS revealed the successful production of cremeomycin as confirmed by comparison to an authentic standard (Figure 2B, Figures S2-3,6). Cremeomycin was not detected in extracts from the wild-type S. lividans TK-64 strain. These results strongly suggest that the cre cluster contains all of the genes necessary for cremeomycin biosynthesis, providing the first experimental link between a set of genes and diazo biosynthesis. Importantly, the S. lividans TK-64 genome does not contain either a cre-type or lom-type putative diazo gene cassette, affording further confirmation that this reactivity is mediated entirely by the cre gene cluster. The success of this heterologous expression therefore narrows the candidate genes encoding diazo-forming enzymes to creABDEM, marking a critical first step towards future mechanistic characterization.
We also identified a second metabolite produced by both S. cremeus and S. lividans TK-64::cre, 2-hydroxy-4-methoxybenzoic acid (2,4-HMBA). The compound was initially discovered during efforts to isolate cremeomycin from S. cremeus, and its identity was established using NMR and MS. Extracts from S. lividans TK-64::cre contained 2,4-HMBA as confirmed by LC-MS and LC-MS/MS comparison with a commercial standard (Figure 2B, Figures S4-5). This compound was not present in extracts of wild-type S. lividans TK-64. We hypothesized that this product results from o-diazoquinone reduction. Indeed, cremeomycin incubated in sterile culture media in the absence of light decomposes to produce 2,4-HMBA over several hours to days (Figure S1).
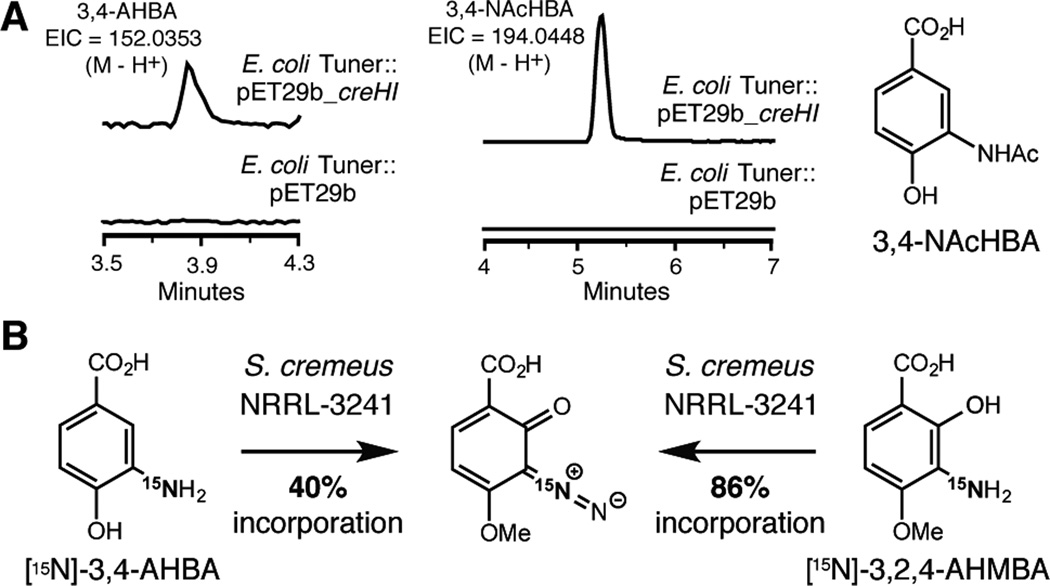
Having confirmed the involvement of the cre cluster in cremeomycin biosynthesis, we next elucidated the order of events in the pathway. We first investigated the functions of CreH/CreI and the role of 3,4-AHBA as a biosynthetic intermediate. We cloned creH and creI together into the expression plasmid pET-29b, which was transformed into Escherichia coli Tuner cells. This strain produced 3,4-AHBA and its N-acetylated derivative 3-acetamido-4-hydroxybenzoic acid, (3,4-NAcHBA) as minor and major products, respectively (Figure 3A). Metabolite identities were confirmed by comparison to authentic standards. These molecules were not detected in an E. coli Tuner strain harboring an empty pET-29b vector. Production of 3,4-AHBA and its N-acetylated derivative is consistent with previous studies of griH and griI[10b] and the known arylamine N-acetyltransferase activity of E. coli.[12] We further established the role of 3,4-AHBA in cremeomycin assembly by feeding 15N-labeled 3,4-AHBA to S. cremeus. Analysis of the isotopic pattern of cremeomycin revealed 40% incorporation of the 15N label, confirming this metabolite is on pathway (Figure 3B & Figure S7). These results also demonstrated that diazo formation proceeds via N–N bond formation in agreement with other diazo-containing natural product biosynthetic pathways.[6]
Figure 3.
3,4-AHBA and 3,2,4-AHMBA are intermediates in cremeomycin biosynthesis. A) Heterologous expression of creH and creI in E. coli affords 3,4-AHBA and 3,4-NAcHBA. B) Stable isotope feeding experiments show incorporation of 15N-labeled 3,4-AHBA and 3,2,4-AHMBA into cremeomycin.
Conversion of 3,4-AHBA to cremeomycin must involve hydroxylation at C2, methylation of the C4 hydroxyl group, and diazotization of the C3 amino group. We obtained information about the potential order of these events from the 15N-3,4-AHBA feeding experiment. The LC-MS data contained a 15N-labeled metabolite with a mass matching that of 3-amino-2-hydroxy-4-methoxybenzoic acid (3,2,4-AHMBA). This molecule could arise from 3,4-AHBA via the actions of monooxygenase CreL and O-methyltransferase CreN. We synthesized and fed a 15N-labeled version of this molecule to S. cremeus. Analysis of extracts by LC-MS revealed 86% incorporation of the 15N label into cremeomycin (Figure 3B & Figure S8), confirming that 3,2,4-AHMBA is a biosynthetic intermediate. These data further demonstrated that hydroxylation and O-methylation precede diazotization in cremeomycin biosynthesis.
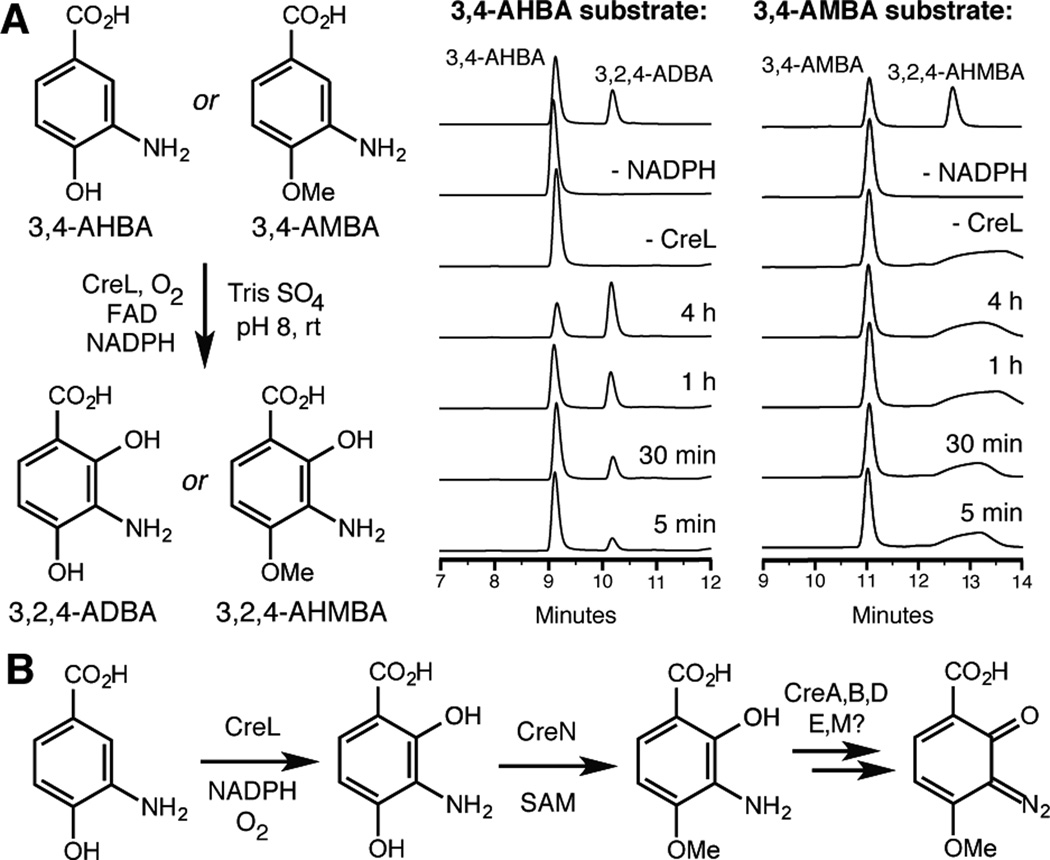
We determined the order of hydroxylation and O-methylation in 3,2,4-AHMBA biosynthesis by characterizing the activities of recombinant CreL and CreN in vitro. Both genes were cloned and expressed in E. coli as C-His6 fusions. CreL-C-His6 purified with bound FAD as determined by LC-MS/MS (Figure S10). Incubation of O-methyltransferase CreN with SAM and either 3,4-AHBA or 3-amino-2,4-dihydroxybenzoic acid (3,2,4-ADBA) and analysis using high performance liquid chromatography (HPLC) showed CreN- and SAM-dependent conversion of both molecules to their respective C4 O-methylated products (Figure S11). The lack of a significant difference in the qualitative rates of O-methylation prevented any insight into the order of events. Conversely, when monooxygenase CreL was incubated with FAD, NADPH, and either 3,4-AHBA or 3-amino-4-methoxybenzoic acid (3,4-AMBA), a new peak was observed by HPLC only in the presence of 3,4-AHBA (Figure 4A). This product was confirmed to be 3,2,4-ADBA by comparison to an authentic standard and its formation was dependent on CreL and NADPH. No conversion of 3,4-AMBA to 3,2,4-AHMBA was observed under any conditions tested. The substrate specificity of CreL revealed that hydroxylation precedes O-methylation in the transformation of 3,4-AHBA to 3,2,4-AHMBA.
Figure 4.
Hydroxylation precedes methylation in the early stages of cremeomycin biosynthesis. A) HPLC assays of flavin-dependent monooxygenase CreL (254 nm) B) Hypothesis for the biosynthesis of cremeomycin from 3,4-AHBA.
In summary, our results establish the order of events in cremeomycin biosynthesis up to the point of diazotization of 3,2,4-AHMBA, which must be catalyzed by one or more of the remaining enzymes (CreABDEM) (Figure 4B). Several of the annotated functions of these enzymes (N-hydroxylation, activation with ATP) are consistent with this assignment, as similar enzymes have been proposed to be involved in the biosynthesis of piperazic acid, a N–N bond containing non-proteinogenic amino acid that is a building block in nonribosomal peptide biosynthesis.[13] Interestingly, CreABDEM are not related to the proposed diazo-forming enzymes from the lomaiviticin and kinamycin pathways. Neither set of putative diazo-forming genes is found in its entirety in the azamerone biosynthetic gene cluster, which contains only a CreM homolog.[6a] Intriguingly, genes encoding CreD and CreE are clustered elsewhere in the genome of the azamerone producer, Streptomyces sp. CNQ-525. The distinct localization of these genes could explain the lack of azamerone production in attempted heterologous expressions.[6a] The presence of distinct biosynthetic machinery in different diazo-synthesizing gene clusters may reflect multiple evolutionary solutions to the problem of installing this reactive functional group. Understanding these different pathways could prove beneficial in efforts to access diazo groups using biocatalysis and synthetic biology. Our characterization of the early steps of cremeomycin biosynthesis now sets the stage for elucidating the enzymatic chemistry of diazo installation. Addressing this important future challenge should enhance our understanding of how Nature generates reactive functionality and reveal new tools for chemical production.
Supplementary Material
Acknowledgments
We thank Wenjun Zhang (University of California at Berkeley) for providing Streptomyces lividans TK-64.
Footnotes
Contributor Information
Abraham J. Waldman, Department of Chemistry and Chemical Biology, Harvard University, 12 Oxford St, Cambridge, MA 02138, United States
Yakov Pechersky, Department of Chemistry and Chemical Biology, Harvard University, 12 Oxford St, Cambridge, MA 02138, United States.
Peng Wang, Department of Chemistry and Chemical Biology, Harvard University, 12 Oxford St, Cambridge, MA 02138, United States.
Jennifer X. Wang, Small Molecule Mass Spectrometry Facility, Faculty of Arts and Sciences Division of Science, Harvard University, 52 Oxford St, Cambridge, MA 02138, United States
Emily P. Balskus, Email: balskus@chemistry.harvard.edu, Department of Chemistry and Chemical Biology, Harvard University, 12 Oxford St, Cambridge, MA 02138, United States.
References
- 1.a) Ye T, Mckervey MA. Chem. Rev. 1994;94:1091–1160. [Google Scholar]; b) Aggarwal VK, de Vicente J, Bonnert RV. J. Org. Chem. 2003;68:5381–5383. doi: 10.1021/jo0268409. [DOI] [PubMed] [Google Scholar]; c) Davies HML, Manning JR. Nature. 2008;451:417–424. doi: 10.1038/nature06485. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Davies HML, Hansen T, Hopper DW, Panaro SA. J. Am. Chem. Soc. 1999;121:6509–6510. [Google Scholar]; e) Johnston JN, Muchalski H, Troyer TL. Angew. Chem. Int. Ed. 2010;49:2290–2298. doi: 10.1002/anie.200904828. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2010;122:2340–2349. [Google Scholar]; f) Antilla JC, Wulff WD. J. Am. Chem. Soc. 1999;121:5099–5100. [Google Scholar]; g) Antos JM, Francis MB. J. Am. Chem. Soc. 2004;126:10256–10257. doi: 10.1021/ja047272c. [DOI] [PubMed] [Google Scholar]; h) Chen Z, Vohidov F, Coughlin JM, Stagg LJ, Arold ST, Ladbury JE, Ball ZT. J. Am. Chem. Soc. 2012;134:10138–10145. doi: 10.1021/ja302284p. [DOI] [PubMed] [Google Scholar]
- 2.Nawrat CC, Moody CJ. Nat. Prod. Rep. 2011;28:1426–1444. doi: 10.1039/c1np00031d. [DOI] [PubMed] [Google Scholar]
- 3.a) Fusari SA, Frohardt RP, Ryder A, Haskell TH, Johannessen DW, Elder CC, Bartz QR. J. Am. Chem. Soc. 1954;76:2878–2881. doi: 10.1038/173072b0. [DOI] [PubMed] [Google Scholar]; b) Dion HW, Fusari SA, Jakubowski ZL, Zora JG, Bartz QR. J. Am. Chem. Soc. 1956;78:3075–3077. [Google Scholar]; c) Rao KV, Brooks SC, Kugelman M, Romano AA. Antibiotics Ann. 1959;60:943. [PubMed] [Google Scholar]
- 4.a) Stock CC, Reilly HC, Buckley SM, Clarke DA, Rhoads CP. Nature. 1954;173:71–72. doi: 10.1038/173071a0. [DOI] [PubMed] [Google Scholar]; b) Ellison RR, Karnofsky DA, Sternberg SS, Murphy ML, Burchenal JH. Cancer. 1954;7:801–814. doi: 10.1002/1097-0142(195407)7:4<801::aid-cncr2820070419>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]; c) Ahluwalia GS, Grem JL, Hao Z, Cooney DA. Pharmac. Ther. 1990;46:243–271. doi: 10.1016/0163-7258(90)90094-i. [DOI] [PubMed] [Google Scholar]; d) Catane R, Von Hoff DD, Glaubiger DL, Muggiaa FM. Cancer Treat. Rep. 1979;63:1033–1038. [PubMed] [Google Scholar]
- 5.Colis LC, Woo CM, Hegan DC, Li Z, Glazer PM, Herzon SB. Nature Chemistry. 2014;6:504–510. doi: 10.1038/nchem.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Winter JM, Jansma AL, Handel TM, Moore BS. Angew. Chem. Int. Ed. 2009;48:767–770. doi: 10.1002/anie.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2009;121:781–784. [Google Scholar]; b) Gould SJ, Melville CR, Cone MC, Chen J, Carney JR. J. Org. Chem. 1997;62:320–324. doi: 10.1021/jo961486y. [DOI] [PubMed] [Google Scholar]
- 7.a) Kersten RD, Lane AL, Nett M, Richter TKS, Duggan BM, Dorrestein PC, Moore BS. ChemBioChem. 2013:955–962. doi: 10.1002/cbic.201300147. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Janso JE, Haltli BA, Eustáquio AS, Kulowski K, Waldman AJ, Zha L, Nakamura H, Bernan VS, He H, Carter GT, Koehn FE. Tetrahedron. 2014;70:4156–4164. doi: 10.1016/j.tet.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Bergy ME, Pyke TR. Cremeomycin and Process for Making. 3350269 US Patent. 1967; b) McGuire JN, Wilson SR, Rinehart KL. J. Antibiot. 1995;48:516–519. doi: 10.7164/antibiotics.48.516. [DOI] [PubMed] [Google Scholar]
- 9.Varley LM, Moody CJ. Synthesis. 2008;22:3601–3604. [Google Scholar]
- 10.a) Ohnishi Y, Furusho Y, Higashi T, Chun H-K, Furihata K, Sakuda S, Horinouchi S. J. Antibiot. 2004;57:218–223. doi: 10.7164/antibiotics.57.218. [DOI] [PubMed] [Google Scholar]; b) Suzuki H, Ohnishi Y, Furusho Y, Sakuda S, Horinouchi S. J. Biol. Chem. 2006:36944–36951. doi: 10.1074/jbc.M608103200. [DOI] [PubMed] [Google Scholar]
- 11.Noguchi A, Kitamura T, Onaka H, Horinouchi S, Ohnishi Y. Nat. Chem. Biol. 2010;6:641–643. doi: 10.1038/nchembio.418. [DOI] [PubMed] [Google Scholar]
- 12.Chang FC, Chung JG. Curr. Microbiol. 1998;36:125–130. doi: 10.1007/pl00006755. [DOI] [PubMed] [Google Scholar]
- 13.Neumann CS, Jiang W, Heemstra JR, Jr, Gontang EA, Kolter R, Walsh CT. ChemBioChem. 2012;13:972–976. doi: 10.1002/cbic.201200054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.