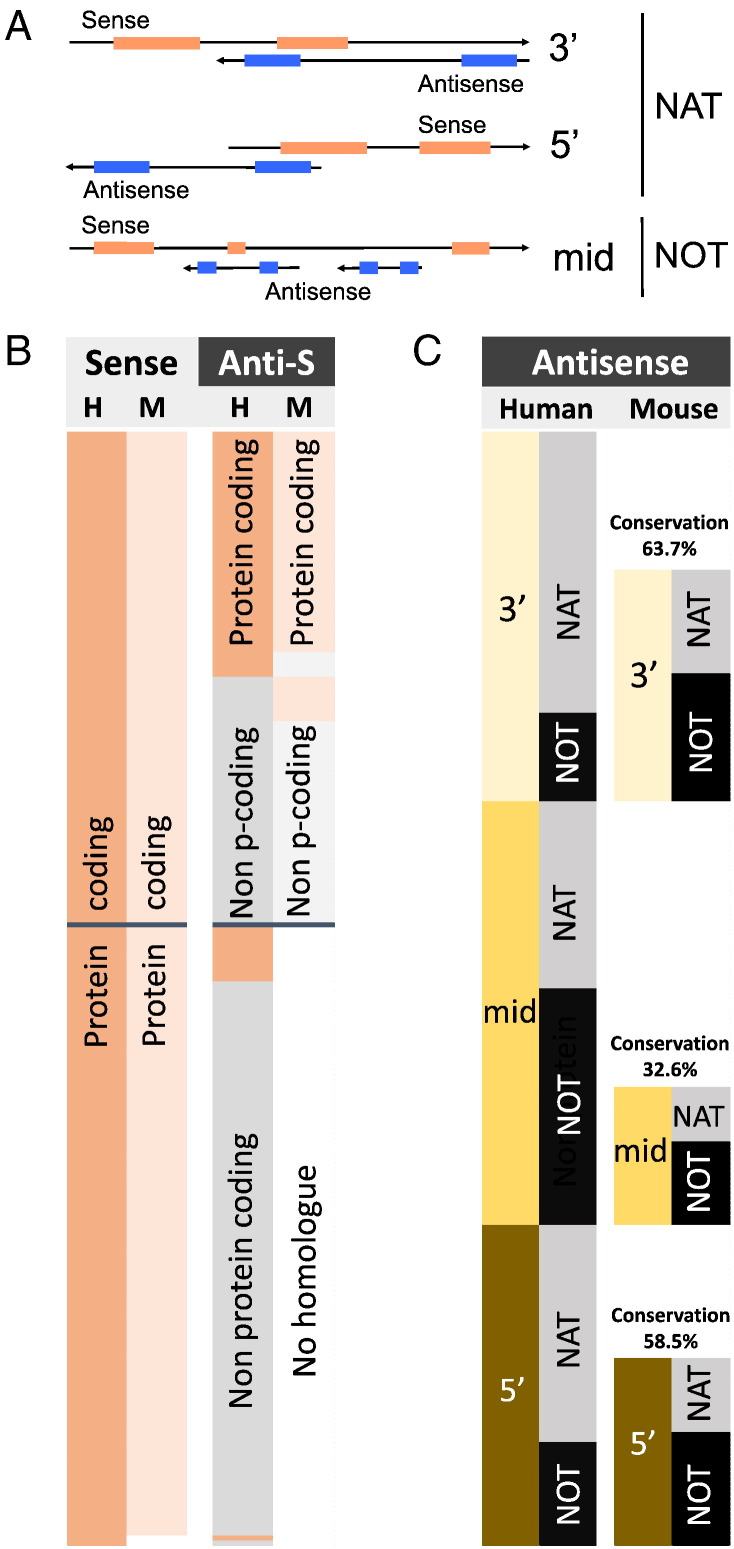
Fig. 3.
Natural antisense transcripts on autosomes of human (H) and mouse (M). A) Classification of sense-antisense overlapping structures including 3′ (tail-to-tail), mid (full overlap) and 5′ (head-to-head). Inclusion into the dataset required complementarity of > 20 nucleotides. B) Expression pattern of sense and antisense transcripts and their phylogenetic conservation. Peach and light peach indicate protein coding potential in human and mouse, respectively. Protein coding sense transcripts are well conserved between human and mouse (left two columns), the antisense transcripts to a much lesser extent (right columns) B) Comparison of the genomic arrangement of bi-directionally transcribed genes in human and mouse. 3′, mid and 5′ indicates the position of complementarity between sense and antisense transcripts. NAT stands for an antisense transcript with complementarity to the sense RNA; NOT indicates a non-overlapping configuration. S = sense, AS = antisense.