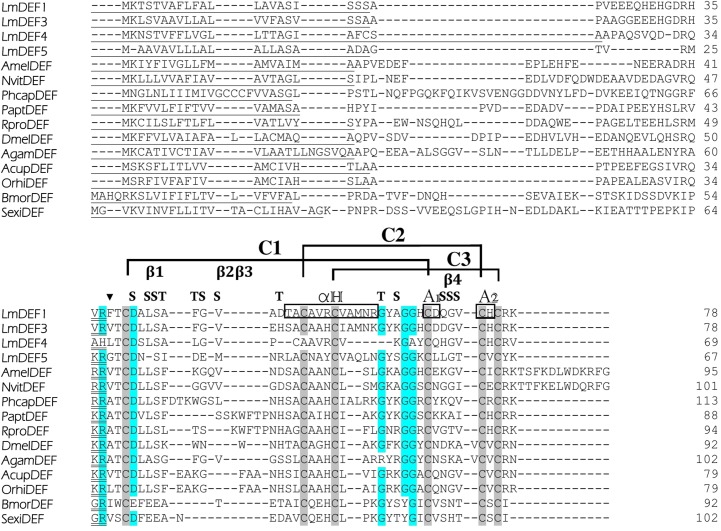
Fig 1. Multiple alignments of newly identified LmDEFs with other insect CSαβ defensins.
Residues conserved in >50% of proteins are shaded. The numbers to the right refer to the position of the last residue of each line. Signal peptides are underlined. Bars indicate gaps to optimize the alignments. The six conserved cysteine residues involved in disulfide bridges are gray shaded. Possible activation peptide cleavage sites are marked with a triangle; enzymatic processing sites (e.g. -KR↓) to release the mature peptides are double underlined. The latter was predicted and/or determined based on various bioinformatical tools (PeptideCutter prediction, the cleaver package, and others) and similarity searches with mature insect defensins from GenBank and reported in the literature. The secondary elements (loop, α-helix, and β-sheet) of insect defensins are indicated as follow: β1, β2, β3, and β4 are regions for potential β turns; A1 and A2 are regions for potential β strands; H is a region for potential α-helix; C1, C2, and C3 are the potential disulfide linkages; S are the regions for bends; T are regions for hydrogen-bonded turns of CSαβ defensins (Following Dassanayake et al. [10]). Accession numbers for the selected insect defensins: AmelDEF, Apis mellifera: [GeneBank: NP_001011616.2]; NvitDEF, Nasonia vitripennis: [GeneBank: NP_001159944.1]; PhcapDEF, Pediculus humanus corporis: [GeneBank: XP_002432619.1]; PaptDEF, Pyrrhocoris apterus: [GeneBank: AGI17576.1]; RproDEF, Rhodnius prolixus [GeneBank: AAO74626.1]; DmelDEF, Drosophila melanogaster [GeneBank: AAO72500.1]; AgamDEF, Anopheles gambiae [GeneBank: ABB00983.1]; AcupDEF, Anomala cuprea [GeneBank: BAD77967.1]; OrhiDEF, Oryctes rhinoceros [GeneBank: BAA36401.1]; BmorDEF, Bombyx mori [GeneBank: BAG48202.1]; SexiDEF, Spodoptera exigua [GeneBank: AEW24427.1].