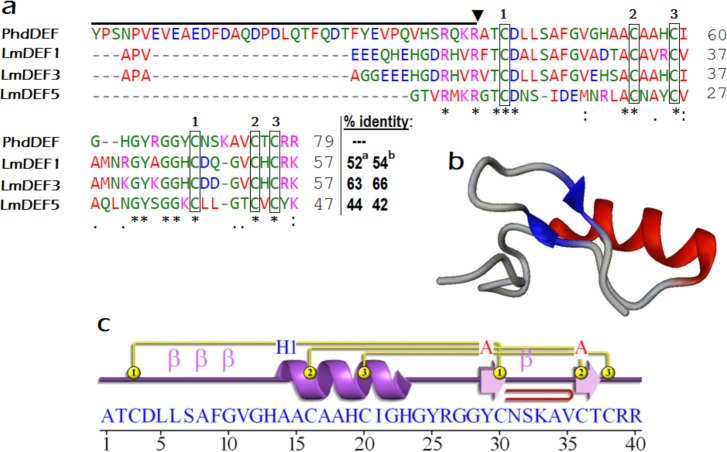
Fig 3. Structural homology of LmDEFs to known antiprotozoal defensin.
a, Alignment of LmDEF1, -3, and -5 with the antiprotozoal Phlebotomus duboscqi defensin (PhdDEF: Boulanger et al. [11]). Percentage identity of LmDEFs to PhdDEF are shown at the end of each individual sequence; a denote the % identity with PhdDEF full peptide excluding the signal peptide, while b is denoting that % with the PhDEF mature active peptide. The amino acid residues are colored according to their physicochemical properties (red: small+ hydrophobic [incl. aromatic -Y]; blue: acidic; magenta: basic–H; green: hydroxyl + sulfhydryl + amine + G). The symbols under the alignment indicate: (*) identical sites; (:) conserved sites; (.) less conserved sites. The boxes indicate the six conserved cysteines; the conserved disulfide bridges are shown by # above these boxes (1–1; 2–2; 3–3). The active peptide cleavage site in PhdDEF is marked with a triangle; while the prodefensin is marked by a line. b, Three-dimensional in silico structure of “active” PhdDEF based on PDB entry 1icaA (defensin A of Protophormia terraenovae) as a template. The homology modeling was carried out with RaptorX; 40(100%) residues were modeled (p-value 9.45e-05). c, The secondary structure elements of PhdDEF predicted by ProFunc server for the purpose of comparison between it and LmDEFs (reported in Fig 2). It comprises 1 sheet, 1 beta hairpin, 1 beta bulge, 2 strands, 1 helix, 4 beta turns, and 3 disulfides.