Figure 1. A divalent, Trx-dependent H3K4me2 chromatin signature marks PREs in Drosophila.
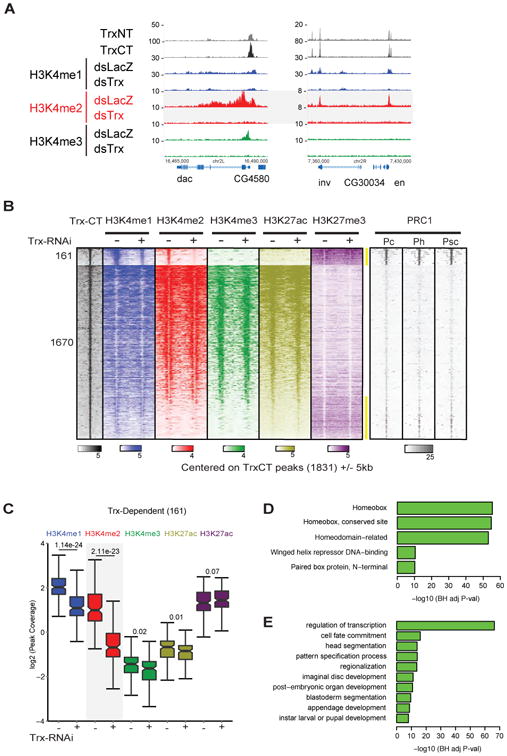
(A) Track examples of PREs at dachshund (dac), invected (inv), and engrailed (en). Both Trx and PRC1 remain bound at the PRE regardless of the gene's transcriptional activity. In the active state, Trx-dependent H3K4-methylation is spread across the gene body (dac), while in the repressed state, H3K4me1/2 is confined to the PRE (inv, en). (B) Analysis of ChIP-seq data after Trx-RNAi. Occupancy levels of Trx-CT and for H3K4me1/2/3, H3K27ac, and H3K27me3 +/- Trx-RNAi. Profiles are centered on Trx-CT occupied peaks (+/- 5kb) and sorted in descending order of H3K4me2 occupancy in WT cells. Group 1 (161 peaks) is distinguished by significantly decreased H3K4me2 after Trx-RNAi and overlap with PRC1-defined PREs. PRC1 ChIP data was obtained from (Enderle et al., 2010). (C) The average occupancy levels per bp within each of the 161 Trx peaks were determined for the five histone modifications and presented in the boxplot. P-values from two-sided student T-tests comparing occupancy +/- Trx-RNAi are shown in the figure. (D,E) For the nearest genes associated with the 161 sites (127 genes), the top 5 Interpro protein domain enrichment results and select top GO biological process results are displayed.
