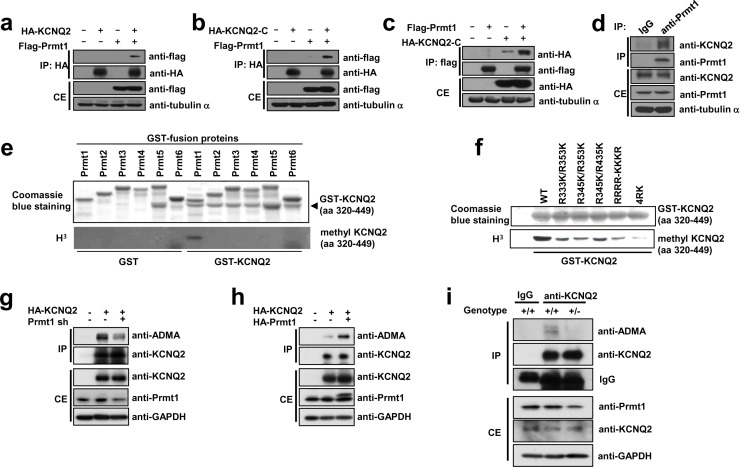
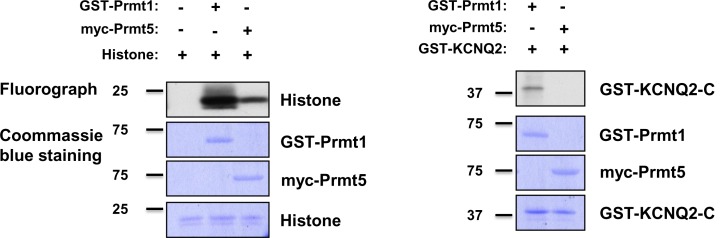
Figure 5. Prmt1 binds to and methylates KCNQ2 at R333, R345, R353, and R435.
(a–c) Immunoblotting analysis showing the physical association of KCNQ2 and Prmt1. HEK293T cells were transfected with expression vectors, as indicated. Whole-cell lysates were immunoprecipitated and immunoblotted by either anti-flag or anti-HA antibody. Representative data from at least three independent experiments are shown. (d) Western blotting analysis showing endogenous interaction of KCNQ2 and Prmt1. Cell lysates from mouse hippocampus were immunoprecipitated with anti-Prmt1 antibody and were immunoblotted with anti-KCNQ2 antibody or anti-Prmt1 antibody. Representative data from at least three independent experiments are shown. (e) In vitro methylation assays with GST-KCNQ2 (amino acids 320–449) and a series of GST-Prmts (1–6) in the presence of [3H]S-adenosylmethionine (SAM). Total amounts of GST-KCNQ2 (arrowhead) and GST-Prmts are shown by Coomassie brilliant blue staining. (f) In vitro methylation assays with GST-Prmt1 together with GST-KCNQ2 (amino acids 320–449) WT, R333K/R353K, R345K/R353K R345K/R435K. R333K/R345K/R353K (RRRR-KKKR), or R333K/R345K/R353K/R435K (4RK) in the presence of [3H]SAM. (g) Immunoblotting analysis showing the decreased asymmetric dimethylation of KCNQ2 in Prmt1 knockdown cells. HEK293T cells were transfected with control or HA-KCNQ2 in combination with control or Prmt1 shRNA expression vectors, followed by immunoprecipitation with KCNQ2 antibodies and immunoblotting with antibodies to ADMA, KCNQ2, Prmt1 and GAPDH. (h) Immunoblotting analysis showing the enhanced asymmetric dimethylation of KCNQ2 in Prmt1 overexpressing cells. HEK293T cells were transfected with control or HA-KCNQ2 in combination with control or Prmt1 expression vectors, followed by immunoprecipitation with KCNQ2 antibodies. (i) Immunoblotting analysis showing decreased asymmetric dimethylation of KCNQ2 in Prmt1+/- brain, compared to the WT control. Brain lysates from Prmt1+/+ and Prmt1+/- mice were immunoprecipitated with anti-KCNQ2 antibodies or control IgG.
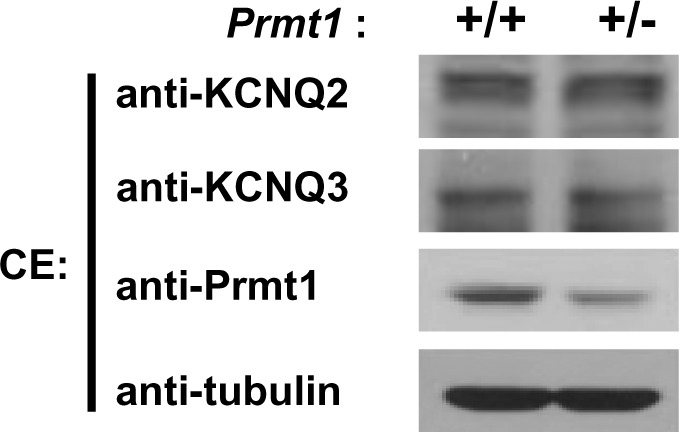
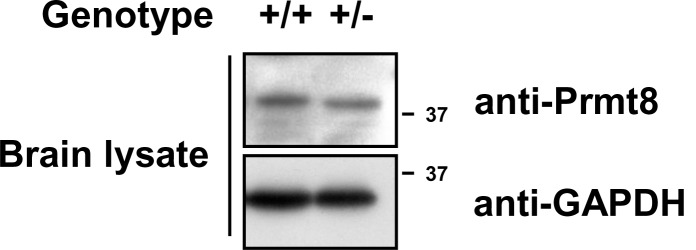
Figure 5—figure supplement 1. Expression of KCNQ2 and KCNQ3 in the hippocampus of WT and Prmt1+/- mice.