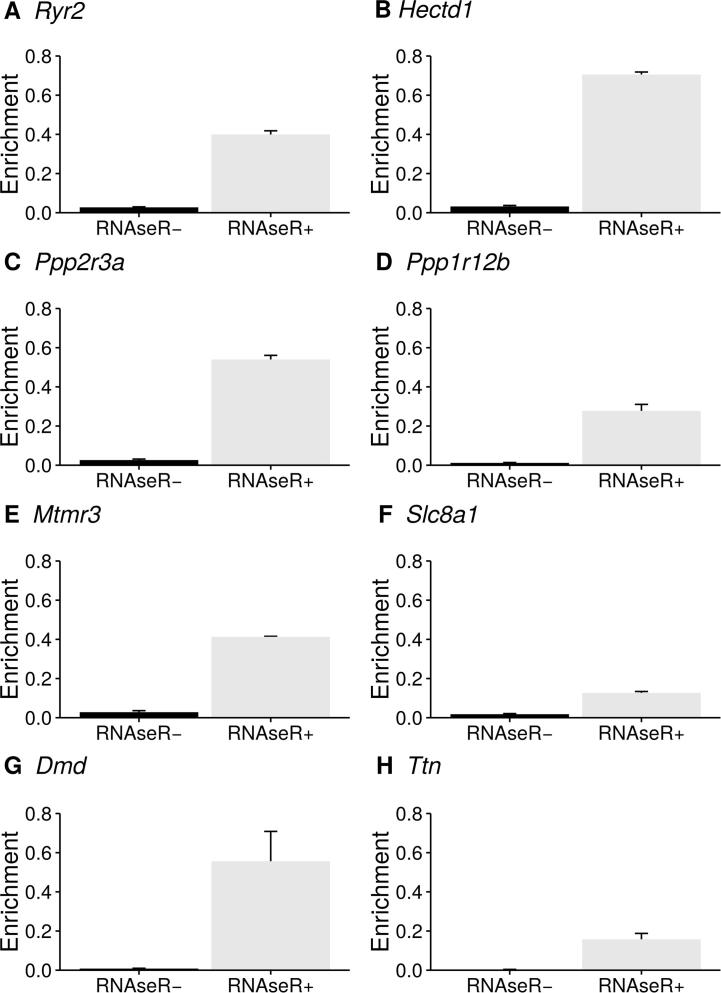
Figure 1.
Comparison of the enrichment for circRNAs in RNase R+ and RNase R− samples
The 8 most significant circRNAs assessed by circTest and linked to certain types of cardiovascular disease are shown (BH corrected P values, FDR 1%). circTest uses the beta binomial distribution to model the data and performs an ANOVA to identify circRNAs that differ in their relative expression between the groups. Due to the implementation of circTest, the 4 time points for the same samples are treated as 4 replicates (n = 4) for RNase R treated (RNase R+) and untreated (RNase R−) samples. Error bars show the standard error of the mean. Therefore, changes in expression at all 4 time points are taken into account when calculating the host gene independence. Ryr2, ryanodine receptor 2; Hectd1, HECT domain-containing E3 ubiquitin protein ligase 1; Ppp2r3a, protein phosphatase 2, regulatory subunit B; Ppp1r12b, protein phosphatase 1, regulatory subunit 12B; MTMR3, myotubularin-related protein 3; Slc8a1, solute carrier family 8 (sodium/calcium exchanger), member 1; Dmd, dystrophin; Ttn, titin.