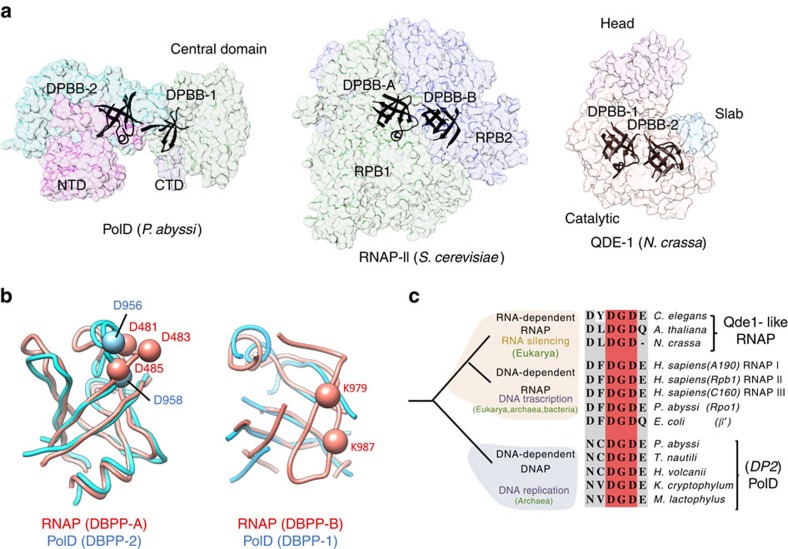
Figure 5. Shared active site architecture between PolD DP2 and ‘two-barrel' RNAPs.
(a) Overview of the conserved ‘two-barrel' catalytic core in PolD DP2, S. cerevisiae RNAP-II (PDBid: 4BBS (ref. 39)) and Neurospora crassa QDE-1 (PDBid: 2J7O (ref. 41)). (b) Superposition of the DPBB subdomains of PolD (blue) and S. cerevisiae RNAP-II (pink). Left: the PolD DPBB-II subdomain is superimposed on the RNAP-II DPBB-A subdomain (Cα r.m.s.d. of 1.72 Å calculated over 73 residues). Cα of the catalytic aspartate residues are shown as spheres. Right: the PolD DPBB-I subdomain is superimposed on the RNAP-II DPBB-B subdomain (Cα r.m.s.d. of 2.21 Å calculated over 42 residues). (c) Possible evolutionary relationship between the DNA-dependent DNAP PolD, DNA-dependent RNAPs and RNA-dependent RNAPs. Conserved catalytic motifs are highlighted in a multi-sequence alignment. The alignment was generated using representative protein with a large sequence diversity to illustrate sequence variability (GI accession number): (i) for RNA-dependent RNAPs Caenorhabditis elegans (392,886,219), Arabidopsis thaliana (42,569,168) and N. crassa (85,091,735); (ii) for DNA-dependent RNAPs Homo sapiens (4,096,591; 119,610,588; 20,159,751), Pyrococcus abyssi (499,169,463) and Escherichia coli (983,454,941); and (iii) for D-family DNAPs P. abyssi (504,648,395), Thermococcus nautili (757,137,858), Haloferax volcanii (490,144,762), Korarchaeum cryptofilum (501,267,152) and Methanosarcina mazei (814,797,709).