FIGURE 5.
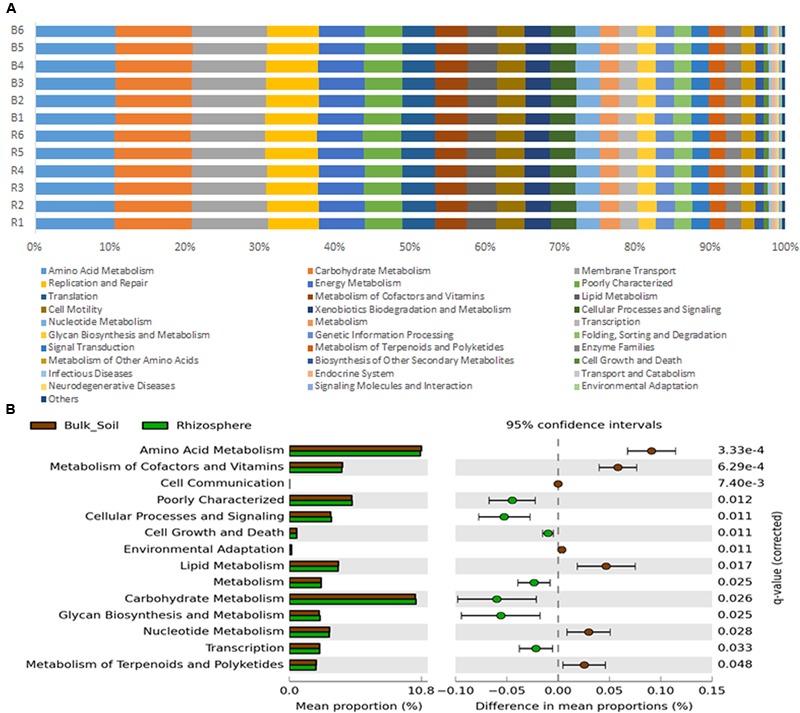
(A) Metagenome predicted functions classified using KEGG level 2 database in PICRUSt software, showing the most abundant functions throughout the 12 samples. (B) Statistical comparison (Welch’s t-test) between the predicted functions abundance on rhizosphere and bulk soil using the Benjamini–Hochberg P-value correction (P < 0.05).
