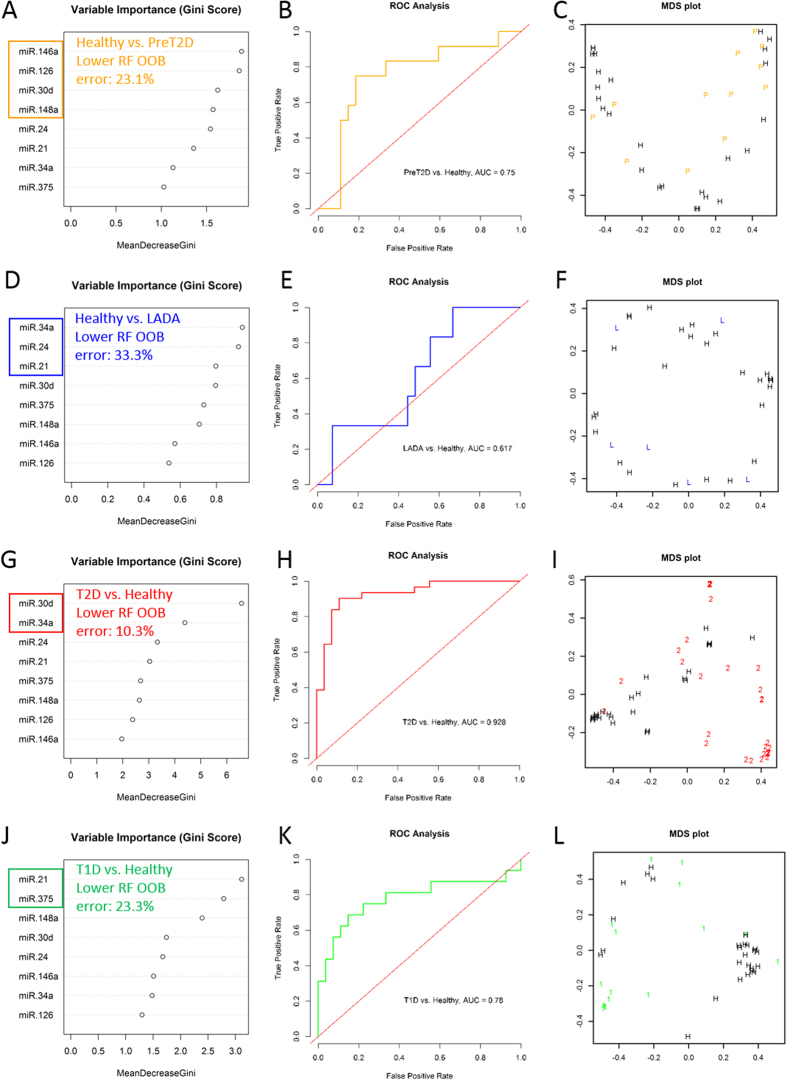
Figure 2. Binary RF classification of diabetes subtypes versus Healthy designation.
Random Forest (RF) classification was performed in the R environment using the randomForest package. Data used was BMI, age, and gender-adjusted logFC for differentially abundant circulating miRNAs and the clinical designation (class label) of each individual [i.e., Healthy, Prediabetes, LADA, T2D, and T1D]. Four sets of binary classifications were conducted, one for each diabetes subtype as compared to Healthy control group. (A–C) shows the results for Prediabetes vs. Healthy classification. (D–F) shows the results for LADA vs. Healthy classification. (G–I) shows the results for T2D vs. Healthy classification. (J–L) shows the results for the T1D vs. Healthy classification. Left panels display the variable importance plot (Gini scores) determined during the initial binary RF classification including all 8 differentially abundant circulating miRNAs. This order of variable importance was used to recursively repeat the RF classification including the top 2, 3, and so forth combinations of miRNAs as predictor variables, and identify the binary classifier with the lower out-of-bag (OOB) estimate of error rate. Outline-colored boxes enclose the combination of miRNAs that generated the classifier with the lower OOB error rate (reported in the top left corner of each left panel graph). The middle panels display the Receiver Operator Characteristic (ROC) Curve generated for sensitivity analysis using the ROCR package. The RF prediction probabilities were used for the generation of the ROCR prediction object. The area under the curve (AUC) is reported as performance measure. The right panels display the multidimensional scaling (MDS) plots for each respective binary RF classification. Color and symbol coding: black and H: Healthy group; orange, P, and PreT2D: Prediabetes group; blue and L: LADA group; red and 2: T2D group; green and 1: T1D group.