Fig. 2.
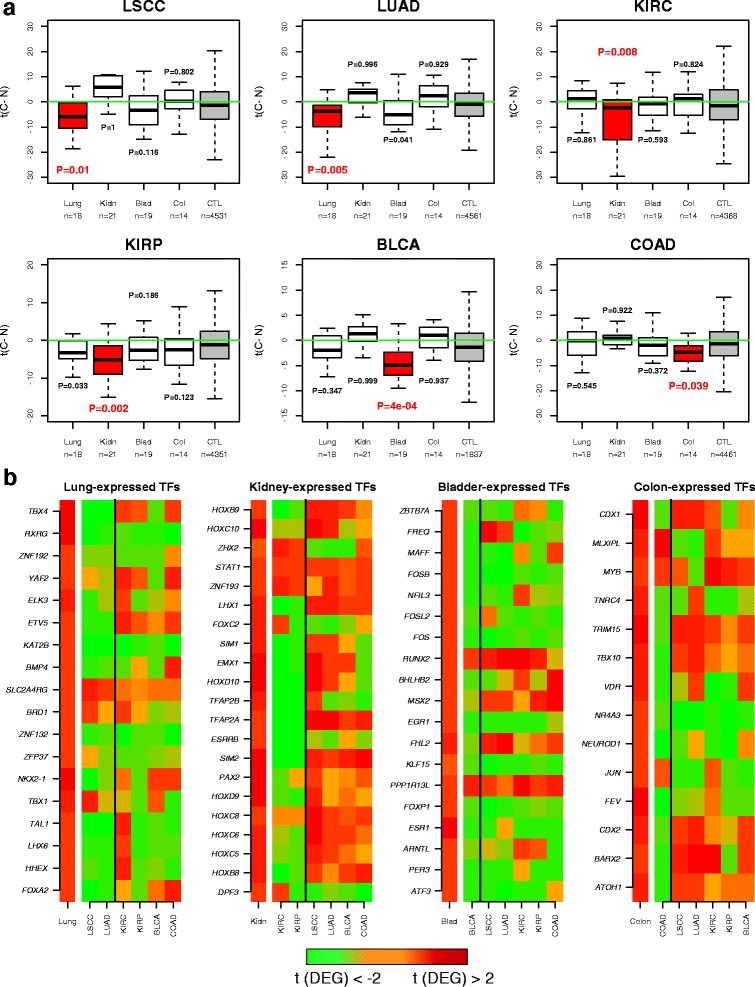
Transcription factors expressed in normal tissue are preferentially silenced in the corresponding cancer type. a Boxplots of t-statistics of differential mRNA expression between cancer and normal tissue (y-axis, t(C − N)) for four sets of “tissue-specific” TFs and a control set of genes (CTL) across six different cancer types, as indicated. LSCC lung squamous cell carcinoma, LUAD lung adenoma carcinoma, KIRC kidney renal clear cell carcinoma, KIRP kidney renal papillary carcinoma, BLCA bladder carcinoma, COAD colon adenoma carcinoma. The five sets of genes being compared are the TFs expressed in the relevant normal tissue (red box), the TFs expressed in other normal tissue types (white boxes) and a set of control (CTL, grey box) non-housekeeping genes which are expressed at a similar level to the TFs expressed in that same normal tissue. P values are from a one-tailed Wilcoxon-rank sum test comparing the t-statistics of each group of TFs with the control (CTL) gene set. We note that negative t-statistics means lower expression in cancer compared with normal. b Heatmaps depicting the dynamics of gene expression changes of the tissue-specific TFs expressed in the normal tissue. t-statistics of differential expression (t(DEG)), are shown between hESCs and normal tissue (the left-most colour heatmap in each panel) and between normal tissue and various cancer types (the right heatmap in each panel), as indicated. We note that the heatmap to the very left in each panel is always red, indicating the overexpression of these TFs in foetal/adult normal tissue compared with hESCs. The heatmap representing the t-statistics of differential expression between normal tissue and the corresponding cancer types are shown to the left of the vertical black line, whereas those for the other unrelated cancer types are shown to the right. There is generally more green (i.e. underexpression) in the cancer types matching the tissue types compared with the other cancer types, in agreement with the data shown in a
