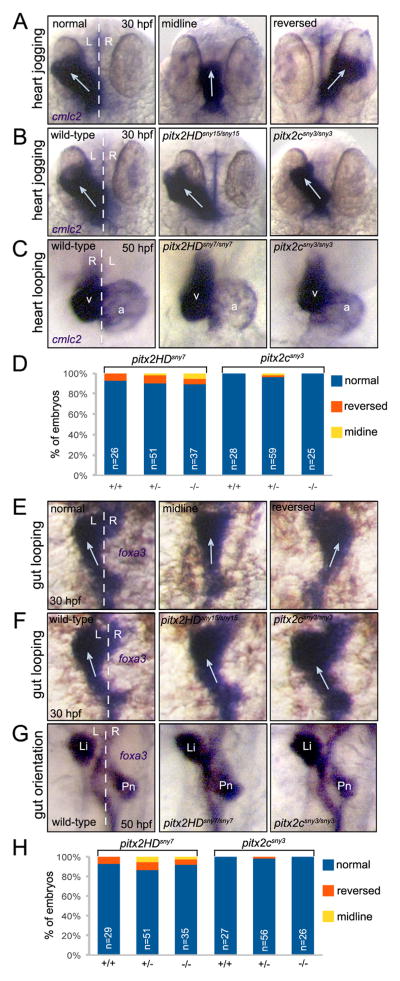
Fig. 6.
Direction of asymmetric heart and gut morphogenesis is normal in pitx2 mutants. (A–C) RNA in situ hybridizations using cmlc2 probes labels the developing heart tube. (A) Selected wild-type embryos that show the potential outcomes of heart jogging at 30 hpf, which include normal left-sided jogging, jogging along the midline and reversed jogging. (B) Representative images of heart jogging in wild-type and pitx2HD and pitx2c mutant embryos. Arrows indicate direction of heart jogging. (C) Normal asymmetric heart looping in wild-type and pitx2HD and pitx2c mutant embryos placed the ventricle (v) to the right of the atrium (a) at 50 hpf. (D) Heart looping direction was scored as either normal, reversed or along the midline for individual embryos at 50 hpf and the genotype (+/+, +/− or −/−) was then determined for each embryo. (E–G) foxa3 in situ hybridizations marked the developing gut tube. (E) Potential outcomes of gut looping at 30 hpf include normal left-sided looping, remaining at the midline and reversed looping. (F) Direction of gut looping was similar in wild-type and pitx2HD and pitx2c mutant embryos. Arrows indicate direction of gut looping. (G) Normal asymmetric placement of the liver (Li) and pancreas (Pn) at 50 hpf. (H) Gut asymmetry was scored at 50 hpf and then each embryo was genotyped. No statistical differences in heart looping (D) or gut asymmetry (H) were identified using single factor one way ANOVA. White dashed lines represent the midline; L=left; R=right; n=number of embryos analyzed.