FIG 7.
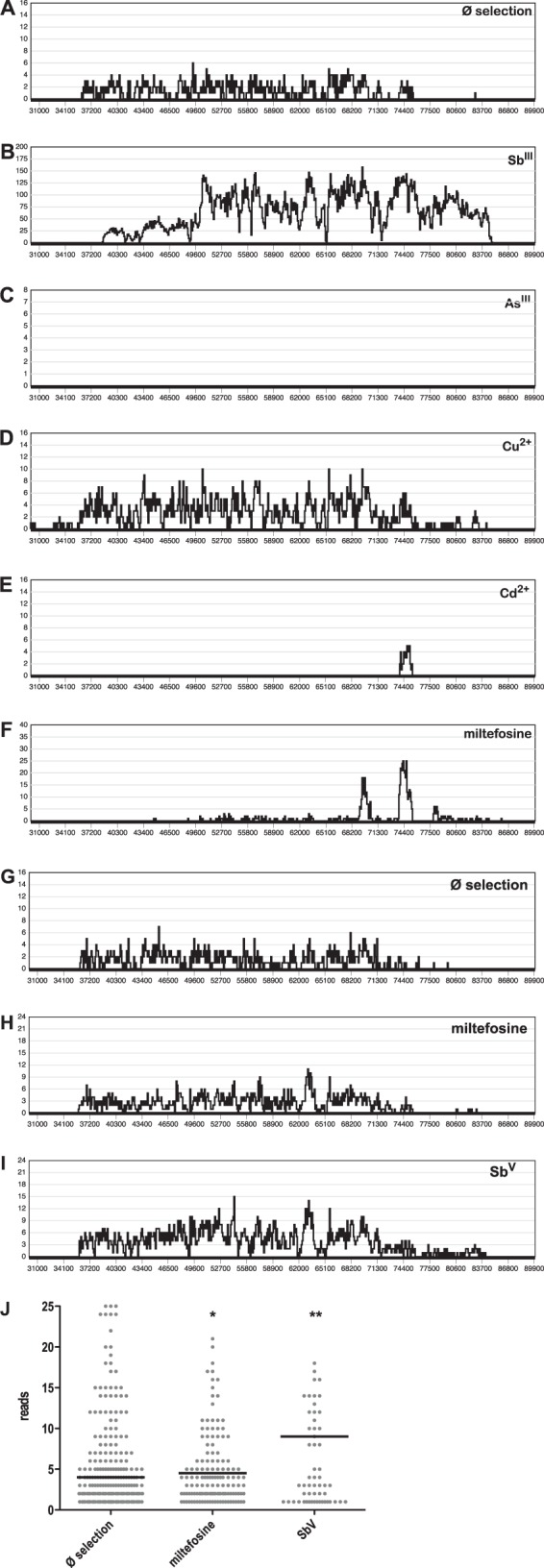
Cos-Seq analysis of L. infantum carrying an L. infantum gDNA cosmid library selected under standard growth conditions (Ø selection) (A) and with antimonyl tartrate (SbIII) (B), sodium arsenite (AsIII) (C), copper acetate (Cu2+) (D), cadmium acetate (Cd2+) (E), and miltefosine (F) at their IC50s. The same transfected population was also used to infect bone marrow-derived macrophages, which were selected by in vitro infection only (G), under miltefosine (H), or under sodium stibogluconate (SbV) (I). In vitro macrophage passages under conditions of drug selection were repeated three times. Cosmid DNA was recovered from all selected populations and subjected to next-generation sequencing. Sequence reads were then aligned with the sequence of L. infantum chromosome 34. In panels A to I, the alignment frequency (y axis) is plotted against chromosomal positions (x axis). (J) Scatter graph analysis of alignment frequencies at positions 36000 to 75000 on chromosome 34 for next-generation sequencing reads from selected intracellular amastigotes (G to I). The black bars represent the median alignment frequencies. *, P < 0.05; **, P < 0.01.
