Fig. (2).
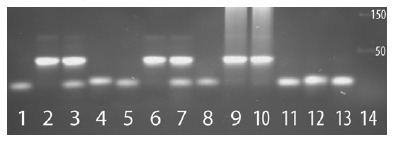
Virus vs. human selenoprotein antisense interactions demonstrated at the DNA level for the Ebola nucleoprotein and HIV nef regions shown in Fig. 1. Target-specific in vitro DNA hybridization of the cognate virus-selenoprotein pairs is shown by gel shift assay using ~40mer synthetic single stranded ssDNA oligos. The matched pairs of oligos from the respective viral and host mRNAs, corresponding to the sequence regions involved in the antisense interactions shown in Fig. 1, were as follows: EBOV nucleoprotein (Enp) vs. TR3, and HIV-1 nef/LTR region (Hnf) vs. TR1. Lanes have either a single oligo that runs as ssDNA (the lowest bands) or an incubated pair of oligos, as follows: 1. Enp; 2. 1:1 Enp+TR3; 3. 2:1 Enp+TR3; 4. TR3; 5. Hnf; 6. 1:1 Hnf+TR1; 7. 2:1 Hnf+TR1; 8. TR1; 9. 1:1 Enp+TR3 plus sheered herring sperm DNA; 10. 1:1 Hnf+TR1 plus herring DNA; 11. 1:1 Enp+TR1; 12. 1:1 Hnf+TR3; 13. 1:1 Hnf+Enp; 14. DNA size markers. The bright bands at the size of ~40mer double stranded dsDNA correspond to the expected hybridizations (Enp+TR3 or Hnf+TR1) at 1:1 (lanes 2,6,9,10) and 2:1 ratios (lanes 3,7). The faint bands above the 50 size marker (e.g. lanes 2,3,6,7) are possibly from trimer formation (e.g. TR3+Enp+TR3, etc.).
