Fig. (3).
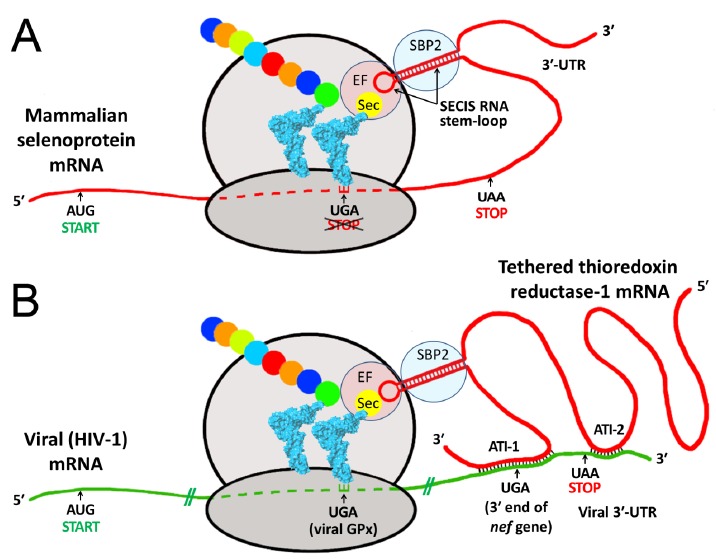
Proposed mechanism of selenocysteine (Sec) incorporation into viral proteins via hijacking of a SECIS element from a tethered host selenoprotein mRNA. Both panels show schematic ribosomes with bound tRNAs (blue), one carrying the rare selenium-containing amino acid Sec, the other a growing peptide chain (colored beads). A. During biosynthesis of mammalian selenoproteins, Sec is inserted at UGA codons, normally a STOP signal for protein synthesis. This mechanism requires a specialized RNA stem-loop structure, the SECIS element, generally located in the 3′ untranslated region (3′-UTR) of the selenoprotein mRNA [4]. By recruiting various protein factors, including SECIS binding protein 2 (SBP2) and elongation factor Sec (EF), the SECIS element enables delivery of tRNASec to the ribosome for Sec incorporation at the UGA codon, preventing it from acting as a stop signal. B. Using HIV-1 as an example, the lower panel shows how a viral mRNA, via antisense tethering interactions (ATI), could hijack a host SECIS element for decoding viral selenoprotein modules, such as the HIV-encoded glutathione peroxidase (viral GPx) [7-8, 13]. ATI-1 is the interaction shown as structure B in Fig. 1, and spans the highly conserved 3′-UGA codon of the nef gene; ATI-2 is a second shorter antisense region, consisting of 13 consecutive Watson-Crick base pairs near the end of the viral mRNA (bases 9111-9123, CAGCUGCUUUUUG). Similarly, in the Ebola nucleoprotein mRNA, less than 350 bases from the ATI shown as structure A in Fig. 1, there is also a secondary ATI region downstream of the conserved 3′-UGA stop codon (bases 2719-2734, CGACAAAUAGCUAACA), with only one mismatch to TR3 over 16 base pairs (A, opposite a G in TR3).
