Abstract
The expression and function of long non-coding RNAs (lncRNAs) in clear cell renal cell carcinoma (ccRCC) remains unclear. The present study measured the expression profiles of three lncRNAs (uc009yby.1, ENST00000514034, and ENST00000450687) using reverse transcription-quantitative polymerase chain reaction, and assessed their signatures in distinguishing ccRCC from matched normal tissues via analysis of receiver operating characteristic (ROC) curves. The expression of uc009yby.1 was inhibited by transfection of renal cells with small interfering RNA, and then the cell proliferation was evaluated by using a Cell Counting Kit-8. The results showed that the expressions of uc009yby.1 and ENST00000514034 were markedly increased in ccRCC compared with the matched normal tissues (P<0.0001 and P=0.0008, respectively), whereas the ENST00000450687 expression was not significantly altered. ROC curves yielded an area under the curve (AUC) value of 0.7000 for uc009yby.1, with sensitivity of 54.29% and specificity of 82.86%; and an AUC value of 0.6627 for ENST00000514034, with sensitivity of 60.00% and specificity of 67.14%. Furthermore, knockdown of uc009yby.1 suppressed renal cell proliferation (Day 0, P=0.7844; Day 1, P=0.0018; Day 2, P=0.0001; Day 3, P<0.000; Day 4, P<0.0001). Taken together, these findings suggest that the expression profiles of uc009yby.1 and ENST00000514034 may serve as novel biomarkers for ccRCC detection, and that uc009yby.1 is strongly associated with renal cell proliferation.
Keywords: long non-coding RNA, clear cell renal cell carcinoma, proliferation, survival
Introduction
Renal cell carcinoma (RCC) accounts for ~3% of adult malignancies (1), resulting in 113,315 mortalities globally in 2008 (2). Clear cell RCC (ccRCC) is the most prevalent subtype of RCC and represents ~70% of all renal tumors (3). The current treatment options for ccRCC comprise chemotherapy, radiotherapy and surgery; however, the only curative treatment for renal cancer is surgery at the time of early diagnosis (4). Metastasis is the major cause of ccRCC-associated mortalities, and around one-third of patients with ccRCC have metastatic tumors at diagnosis (5). Thus, it is crucial to identify predictive biomarkers for early detection of ccRCC.
Numerous studies have indicated that 70–90% of the human genome transcribes RNA products, but only 2% of the total genome encodes protein-coding genes (6). Long non-coding RNAs (lncRNAs) are a large class of non-coding RNAs of 200–100,000 nucleotides in length which can interact with DNA, RNA and/or proteins to regulate gene expression or affect protein activity in human disease, including RCC (7). For example, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) interacts with enhancer of zeste homolog 2 and microRNA-205 to suppress the E-cadherin expression in RCC cells, suggesting that MALAT1 may offer a novel theranostic marker for RCC (8). An increasing number of studies have also demonstrated that certain lncRNAs are strongly associated with patient survival and early pathological changes in ccRCC (9,10). Recently, a large number of lncRNAs that have aberrant expression in ccRCC were screened via lncRNA microarray in several studies (11–13); however, the expression of these lncRNAs have not been further confirmed and their functions in ccRCC cells have not been explored. In the current study, the expression profiles of three lncRNAs (uc009yby.1, ENST00000514034, and ENST00000450687) in ccRCC tissues were detected, and their efficacy in detecting ccRCC was analyzed. Furthermore, the role of uc009yby.1 in renal carcinoma cells was investigated.
Materials and methods
Patient samples
In total, 70 patients with ccRCC were included in the present study between 2009 and 2014 at Wuhan General Hospital (Wuhan, China). All samples were collected from patients who had undergone nephrectomy prior to any treatment, and were histologically confirmed (Table I). The study protocols were approved by the Ethics Review Board of the hospital and all enrolled patients provided written informed consent for tissue donation and publication of findings. The follow-up of patients was performed from the date of surgery until the time of mortality or the end of the study period in November 2013. Among the 70 patients, the follow-up of 5 patients was lost.
Table I.
Clinical features of the included patients with clear cell renal cell carcinoma (n=70).
| uc009yby.1 expression | ENST00000514034 expression | ||||||
|---|---|---|---|---|---|---|---|
| Variable | Cases, n | Low, n | High, n | P-value | Low, n | High, n | P-value |
| Gender | 0.809 | 0.809 | |||||
| Male | 40 | 21 | 19 | 21 | 19 | ||
| Female | 30 | 14 | 16 | 14 | 16 | ||
| Age, years | 0.811 | 0.811 | |||||
| Median | 56a | ||||||
| ≥55 | 36 | 19 | 17 | 17 | 19 | ||
| <55 | 34 | 16 | 18 | 18 | 16 | ||
| Tumor diameter, cm | 0.030 | 0.472 | |||||
| ≥4 | 38 | 14 | 24 | 17 | 21 | ||
| <4 | 32 | 21 | 11 | 28 | 14 | ||
| AJCC stage | 0.025 | 0.216 | |||||
| I/II | 26 | 18 | 8 | 10 | 16 | ||
| III/V | 44 | 17 | 27 | 25 | 19 | ||
Range, 35–78 years. P-values were calculated by two-sided χ2 test; values in bold indicate statistical significant (P<0.05). AJCC, American Joint Committee on Cancer.
Cell culture
The RCC cell lines 786-O and ACHN, and the normal human proximal tubule epithelial cell line HK-2 were purchased from the American Type Culture Collection (Manassas, VA, USA). 786-O and ACHN were cultured in HyClone Dulbecco's modified Eagle's medium and minimal essential medium (GE Healthcare Life Sciences, Logan, UT, USA), respectively, and HK-2 was cultured in HyClone KSFM medium (GE Healthcare Life Sciences), each supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and incubated at 37°C in 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from each of the RCC tissues or cell lines using Invitrogen TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The concentration and A260/280 ratio of total RNAs were measured using a NanoVue Plus spectrophotometer (GE Healthcare Life Sciences, Chalfont, UK). Complementary DNA was synthesized from 1 µg of total RNA using a PrimeScript RT Reagent Kit (Takara Biotechnology Co., Ltd., Dalian, China), from which the genomic DNA was removed prior to RT assay. SYBR Green Real-Time PCR Master Mix kits (Takara Biotechnology Co., Ltd.) were used to generate the DNA products in an Applied Biosystems 7500 Fluorescent Quantitative PCR System (Thermo Fisher Scientific, Inc.). A total of 20 µl of the reaction mixtures containing 2 µl of cDNA template were subjected to PCR as follows: Incubation at 95°C for 1 min; followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec. The lncRNA expressions were normalized from the quantification cycle (Cq) values of the lncRNAs relative to β-actin using the 2−ΔΔCq method (14). The primers sequences use in the present study are presented in Table II.
Table II.
Primer sequences.
| Primer | Sequence | Product size, bp |
|---|---|---|
| uc009yby.1 sense | 5′-GGAAGGCTGAGGCAAGAGAATCG-3′ | 243 |
| uc009yby.1 antisense | 5′-GCTGCTGCTGGCTCTGATGAAG-3′ | |
| ENST00000514034 sense | 5′-GGACAGTTGGAACACCTCTTCTCAG-3′ | 216 |
| ENST00000514034 antisense | 5′-GCCAGAGTGAAGTGACGGAGGA-3′ | |
| ENST00000450687 sense | 5′-CTGGACTTGCAGCAGATGACTTCA-3′ | 130 |
| ENST00000450687 antisense | 5′-CTGGAGGGACTGGAGGACTTGTT-3′ | |
| β-actin sense | 5′-GCACTCTTCCAGCCTTCCTTCCT-3′ | 122 |
| β-actin antisense | 5′-GACAGCACTGTGTTGGCGTACA-3′ |
Transfection of small interfering RNA (siRNA)
The siRNAs for uc009vby.1 (si-uc009yby.1) were designed and synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences of siRNAs were as follows: siRNA1, TGTGGCATGACCTGGGTTGCATCTGGCATGGCCC; and siRNA2, TAGCTCTGCAGGTGTGGACCCCAAGACCACACACAG. Transfection of si-uc009yby.1 and the negative control (provided with the kit) was performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol.
Cell proliferation assay
786-O and ACHN cells were seeded in 96-well plates with a density of 104 cells per well after the cells had been transfected with si-uc009yby.1 (100 nM) or negative control (100 nM), respectively. After 1, 2, 3, 4 or 5 days, cell proliferation was analyzed using a Cell Counting Kit 8 (Beyotime Institute of Biotechnology, Haimen, China) according to the manufacturer's protocol.
Statistical analysis
A Mann-Whitney test was used to compare the differences in lncRNA expression between tissues. Differences between groups were analyzed using a two-tailed Student's t-test. The Youden index was used to select the optimal cutoff value. Survival curves were estimated by the Kaplan-Meier method and assessed by log-rank test. P<0.05 was considered to indicate statistically significant differences. All statistical analyses were performed using SPSS version 17.0 software (SPSS, Inc., Chicago, IL, USA).
Results
Expression levels of uc009yby.1, ENST00000514034 and ENST00000450687 are frequently increased in ccRCC tissues
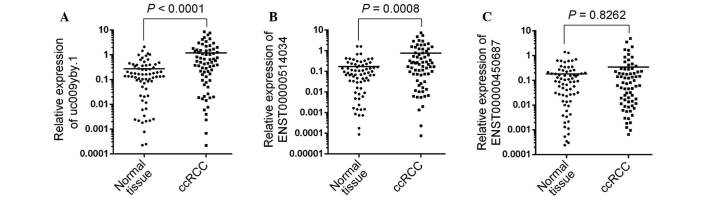
A lncRNA microarray revealed that a large number of lncRNAs had aberrant expression in ccRCC tissues (12). Among the top three upregulated lncRNAs, three lncRNAs (uc009yby.1, ENST00000514034 and ENST00000450687) had not been confirmed on a large scale via RT-qPCR. To evaluate the three lncRNAs expressions in ccRCC, their expression in RCC and matched normal tissues from 70 patients with ccRCC was analyzed. The results showed that the expression levels of uc009yby.1 and ENST00000514034 were markedly increased in RCC tumor tissues compared to the matched normal tissues (P<0.0001 and P=0.0008, respectively) whereas the ENST00000450687 expression was not significantly altered (Fig. 1).
Figure 1.
Expression profiles of lncRNA uc009yby.1, ENST00000514034 and ENST00000450687. Scatter dot plots show the expression levels of (A) uc009yby.1, (B) ENST00000514034 and (C) ENST00000450687 in the tumor tissues and the matched normal tissues of patients with ccRCC (n=70). The lines inside the scatter dot plots denote the median values. lncRNA, long non-coding RNA; ccRCC, clear cell renal cell carcinoma.
Expression profiles of uc009yby.1 and ENST00000514034 serve as biomarkers for ccRCC detection
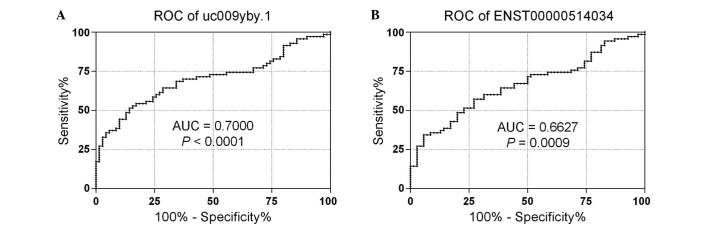
To evaluate the ability of uc009yby.1 and ENST00000514034 on distinguishing ccRCC from normal tissues, receiver operating characteristic (ROC) curves were used to analyze the efficacy of expression profiles of the two lncRNAs in detecting ccRCCs. ROC curves yielded an area under the curve (AUC) of 0.7000 (95% CI, 0.6117–0.7883) for uc009yby.1, with sensitivity of 54.29% and specificity of 82.86%; and an AUC of 0.6627 (95% CI, 0.5721–0.7532) for ENST00000514034, with sensitivity of 60.00% and specificity of 67.14% (Fig. 2). These results suggested that uc009yby.1 and ENST00000514034 may be valuable biomarkers for ccRCC detection.
Figure 2.
ROC curve analyses of the expression profiles of (A) uc009yby.1 and (B) ENST00000514034 in distinguishing ccRCC from matched normal tissues. The interval between the 5th and 95th percentiles denotes the confidence level. ROC, receiver operating characteristic; ccRCC clear cell renal cell carcinoma; AUC area under the curve.
Expression levels of uc009yby.1 and ENST00000514034 are associated with ccRCC patient survival
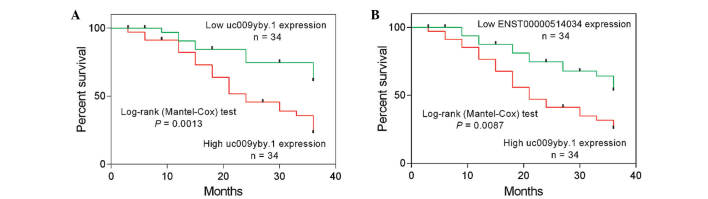
The follow-up of 70 patients with ccRCC was performed, from which 2 patients were lost to follow-up. The expression levels of uc009yby.1 and ENST00000514034 were used to conduct Kaplan-Meier analyses of patient survival. The 68 ccRCC samples were divided into high (above the median) and low (below the median) uc009yby.1 expression groups (n=34 per group), and high (above the median) and low (below the median) ENST00000514034 expression groups (n=34 per group), based on the median value of the two lncRNA levels. As shown in Fig. 3, patients with high uc009yby.1 expression had markedly poorer overall survival times compared to those with low uc009yby.1 expression (P=0.0013), and patients with high ENST00000514034 expression had significantly poorer overall survival times compared to those with low ENST00000514034 expression (P=0.0087). These findings suggest that uc009yby.1 and ENST00000514034 may be used to predict ccRCC patient survival.
Figure 3.
Patient survival analyses. ccRCC patients with high expression of (A) uc009yby.1 and (B) ENST00000514034 had decreased overall survival times compared to patients with low expression of uc009yby.1 and ENST00000514034, respectively.
uc009yby.1 expression is repressed by siRNA in 786-O and ACHN cells
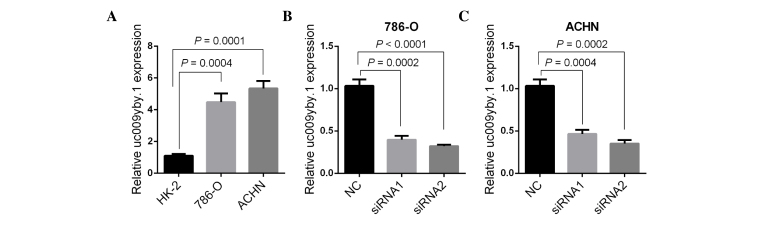
As uc009yby.1 had a more significant effect with regard to ccRCC detection, its function was investigated further. First, uc009yby.1 expression was measured in three cell lines (HK-2, 786-O and ACHN). The results revealed that the uc009yby.1 expression was significantly higher in 786-O and ACHN cells than in HK-2 cells (P=0.0004 and P=0.0001, respectively) (Fig. 4A). siRNA1 and siRNA2 were synthesized and transfected into 786-O and ACHN cells, respectively. Quantification analysis revealed that siRNA1 and siRNA2 each significantly knocked down uc009yby.1 expression compared with the negative control-transfected cells (P=0.0002 and P<0.0001 in 786-O cells, P=0.0004 and P=0.0002 in ACHN cells) (Fig. 4B and C).
Figure 4.
Knockdown of uc009yby.1 by siRNA in renal cells. (A) uc009yby.1 expression was increased in the RCC cell lines 786-O and ACHN compared with the normal renal epithelial cell line HK-2. (B) siRNA1 (100 nM) or siRNA2 (100 nM) was transfected into 786-O cells and inhibited uc009yby.1 expression. (C) ACHN cells transfected with siRNA1 (100 nM) or siRNA2 (100 nM) showed lower uc009yby.1 expression compared with ACHN cells transfected with NC (100 nM). **P<0.01. siRNA, small interfering RNA; NC, negative control.
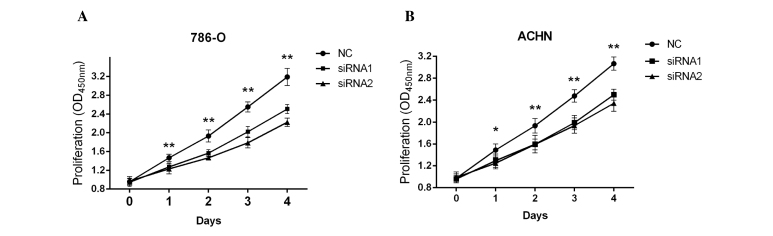
uc009yby.1 facilitates the proliferation of 786-O and ACHN cells
The effect of uc009yby.1 on cell growth of 786-O and ACHN cells was investigated. Following the transfection of 786-O and ACHN cells with negative control and siRNAs, the cell proliferation was analyzed. As shown in Fig. 5, siRNA1 and siRNA2 of uc009yby.1 markedly suppressed the proliferation of 786-O and ACHN cells, respectively. These results suggest that uc009yby.1 plays an important role in ccRCC development.
Figure 5.
Analysis of cell proliferation after renal cells were transfected with siRNAs. (A) siRNA1 (100 nM) and siRNA2 (100 nM) for uc009yby.1 were transfected into 786-O cells and suppressed cell proliferation compared with NC-transfected cells. siRNA1: Day 0, P=0.7844; day 1, P=0.0018; day 2, P=0.0001; day 3, P<0.0001; and day 4, P<0.0001. siRNA2: Day 0, P=0.7592; day 1, P=0.0015; day 2, P<0.0001; day 3, P<0.0001; day 4, P<0.0001. (B) ACHN cells transfected with siRNA1 (100 nM) and siRNA2 (100 nM) for uc009yby.1 revealed attenuated cell proliferation. siRNA1: Day 0, P=0.7289; day 1, P=0.0157; day 2, P=0.0026; day 3, P<0.0001; day 4, P<0.0001. siRNA2: Day 0, P=0.7926; day 1, P=0.0031; day 2, P<0.0005; day 3, P<0.0001; day 4, P<0.0001. siRNA, small interfering RNA; OD, optical density; NC, negative control. *P<0.05; **P<0.001.
Discussion
The current study demonstrated that the expression levels of two lncRNAs (uc009yby.1 and ENST00000514034) were frequently elevated in 70 ccRCC tissues relative to 70 matched normal tissues. This indicates that the expression profiles of the two lncRNAs may serve as novel biomarkers for ccRCC detection. Furthermore, the function of uc009yby.1 in renal cells was investigated using two siRNAs to inhibit uc009yby.1 expression, revealing that this lncRNA could enhance renal cell proliferation; this suggests that uc009yby.1 may be a novel therapeutic target for ccRCC.
Numerous studies have indicated that certain lncRNAs have aberrant expression profiles in various kinds of tumors, such as gastric (15–17), colon (18,19), esophageal (20–22) and liver (23) cancers. Yu et al (12) recently reported on a class of lncRNAs that are aberrantly expressed in ccRCC cells, in which multiple lncRNAs were upregulated. The top five upregulated lncRNAs were ENST00000456816, X91348, uc009yby.1, ENST00000514034 and ENST00000450687. Consistently with this study, the current results confirmed the increased expression of uc009yby.1 and ENST00000514034 in ccRCC tissue samples from patients using RT-qPCR on a large scale; however, the ENST00000514034 expression was not significantly changed, suggesting that the results generated from the lncRNA microarray were false positive and should be further investigated.
Several studies have indicated that lncRNA expression profiles may act as biomarkers for diagnosis of ccRCC; lncRNA MALAT1, for example, was reported to be associated with tumor progression and poor prognosis in ccRCC (9), and lncRNA CADM1-AS1 was associated with poor prognosis in patients with ccRCC. Consistent with these findings, the current data revealed that the expression profiles of uc009yby.1 and ENST00000514034 had high sensitivity and specificity in distinguishing ccRCC from normal tissues. Furthermore, ccRCC patients with high uc009yby.1 and/or ENST00000514034 expression had poorer survival times. This suggested that the signatures of uc009yby.1 and ENST00000514034 may serve as novel biomarkers for the prognosis and diagnosis of ccRCC.
Increasing numbers of studies have demonstrated that lncRNAs are of crucial function in tumor cells, in which they can interact with histones and regulate expression of certain genes, such as Xist (24–27). Loss or gain of lncRNAs could result in the disorder of cell metabolism. Li et al (21) reported that upregulation of lncRNA POU3F3 promoted DNA methylation and enhanced cell proliferation in esophageal squamous cell carcinoma cells; and when POU3F3 expression was knocked down by siRNAs, cell proliferation was significantly suppressed. Recently, Yao et al (10) demonstrated that lncRNA CADM1-AS1 was significantly decreased in ccRCC, and that downregulation of CADM1-AS1 by siRNAs enhanced cell proliferation and migration, and inhibited cell apoptosis in 786-O cells. In the present study, a novel lncRNA, uc009yby.1, was found to be increased in ccRCC, and downregulation of uc009yby.1 by siRNAs could repress cell proliferation in 786-O and ACHN cells, strongly suggesting that uc009yby.1 could regulate renal cell proliferation.
Recently, several studies have demonstrated that lncRNAs are stably presented in the blood of cancer patients, such as lncRNA POU3F3 (28) and lncRNA TapSaki (29). Appearance of circulating lncRNA highlights the potential for noninvasive biomarkers derived from lncRNA expression profiles for prognosis and diagnosis of cancer, as these biomarkers have high sensitivity and specificity. However, in the current study, plasma and serum samples were not collected from patients with ccRCCs to evaluate whether these lncRNA signatures were efficient for diagnosis and prognosis of ccRCC. Whilst the present data revealed that uc009yby.1 could modulate the proliferation of renal cells, the regulatory mechanism remains largely unknown. In future, it will be necessary to expound the regulatory mechanism by which uc009yby.1 contributes to cell proliferation.
Taken together, the results of the present study indicate that the expression of uc009yby.1 and ENST00000514034 is increased in ccRCC tissues compared to matched normal tissues, and these signature may serve as novel biomarkers for ccRCC detection. Furthermore, downregulation of uc009yby.1 is able to suppress renal cell proliferation, suggesting that uc009yby.1 may be a novel therapeutic target for ccRCC.
References
- 1.Chow WH, Dong LM, Devesa SS. Epidemiology and risk factors for kidney cancer. Nat Rev Urol. 2010;7:245–257. doi: 10.1038/nrurol.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Martel CL, Lara PN. Renal cell carcinoma: Current status and future directions. Crit Rev Oncol Hematol. 2003;45:177–190. doi: 10.1016/S1040-8428(02)00076-8. [DOI] [PubMed] [Google Scholar]
- 4.Russo P. Renal cell carcinoma: Presentation, staging, and surgical treatment. Semin Oncol. 2000;27:160–176. [PubMed] [Google Scholar]
- 5.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 6.ENCODE Project Consortium: An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhai W, Sun Y, Jiang M, Wang M, Gasiewicz TA, Zheng J, Chang C. Differential regulation of LncRNA-SARCC suppresses VHL-mutant RCC cell proliferation yet promotes VHL-normal RCC cell proliferation via modulating androgen receptor/HIF-2α/C-MYC axis under hypoxia. Oncogene. 2016 doi: 10.1038/onc.2016.19. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 8.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947–2955. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 10.Yao J, Chen Y, Wang Y, Liu S, Yuan X, Pan F, Geng P. Decreased expression of a novel lncRNA CADM1-AS1 is associated with poor prognosis in patients with clear cell renal cell carcinomas. Int J Clin Exp Pathol. 2014;7:2758–2767. [PMC free article] [PubMed] [Google Scholar]
- 11.Qin C, Han Z, Qian J, Bao M, Li P, Ju X, Zhang S, Zhang L, Li S, Cao Q, et al. Expression pattern of long non-coding RNAs in renal cell carcinoma revealed by microarray. PLoS One. 2014;9:e99372. doi: 10.1371/journal.pone.0099372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu G, Yao W, Wang J, Ma X, Xiao W, Li H, Xia D, Yang Y, Deng K, Xiao H, et al. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS One. 2012;7:e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fachel AA, Tahira AC, Vilella-Arias SA, Maracaja-Coutinho V, Gimba ER, Vignal GM, Campos FS, Reis EM, Verjovski-Almeida S. Expression analysis and in silico characterization of intronic long noncoding RNAs in renal cell carcinoma: Emerging functional associations. Mol Cancer. 2013;12:140. doi: 10.1186/1476-4598-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen JF. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Zhang D, Wu K, Zhao Q, Nie Y, Fan D. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol. 2014;34:3182–3193. doi: 10.1128/MCB.01580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X, He Y, Xi B, Zheng J, Zeng X, Cai Q, Ouyang Y, Wang C, Zhou X, Huang H, et al. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS one. 2013;8:e81437. doi: 10.1371/journal.pone.0081437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang CM, Wu QQ, Li SQ, Chen FJ, Tuo L, Xie HW, Tong YS, Ji L, Zhou GZ, Cao G, et al. Upregulation of the long non-coding RNA PlncRNA-1 promotes esophageal squamous carcinoma cell proliferation and correlates with advanced clinical stage. Dig Dis Sci. 2014;59:591–597. doi: 10.1007/s10620-013-2956-7. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, Wang S, Zhou F, Shi S, Feng X, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63:1700–1710. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang B, Li BS, Yang SM. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360:119–124. doi: 10.1016/j.canlet.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 24.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt AM, Chang HY. Gene regulation: Long RNAs wire up cancer growth. Nature. 2013;500:536–537. doi: 10.1038/nature12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong YS, Wang XW, Zhou XL, Liu ZH, Yang TX, Shi WH, Xie HW, Lv J, Wu QQ, Cao XF. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. doi: 10.1186/1476-4598-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenzen JM, Schauerte C, Kielstein JT, Hübner A, Martino F, Fiedler J, Gupta SK, Faulhaber-Walter R, Kumarswamy R, Hafer C, et al. Circulating long noncoding RNATapSaki is a predictor of mortality in critically ill patients with acute kidney injury. Clin Chem. 2015;61:191–201. doi: 10.1373/clinchem.2014.230359. [DOI] [PubMed] [Google Scholar]