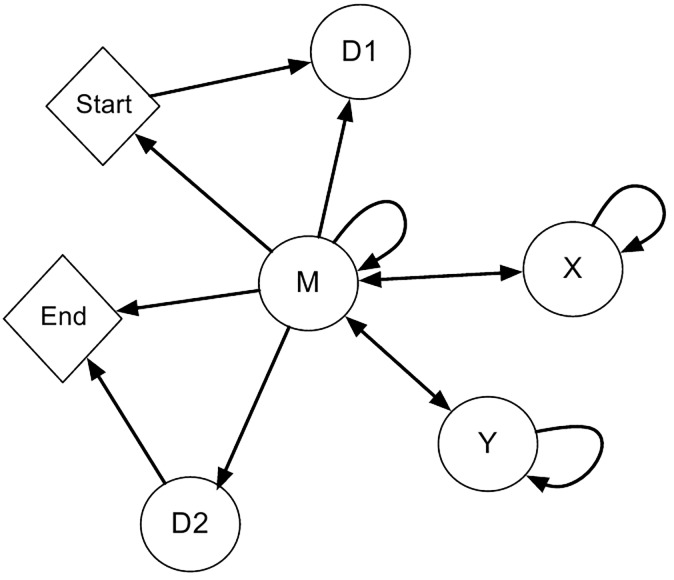
Fig 1. An Illustration of our paired HMM.
Our model is tailored for semi-global alignment with seven states: start, D1, D2, M, X, Y and end. State M represents a match between an amino acid pair in S(6, 1′) and a target RNA nucleotide in an RNA transcript. States D1, D2 and X all represent a gap on the S6, 1′ side of the alignment. State Y represents a gap in the RNA sequence. D1 represents a gap in S(6, 1′) before the occurrence of a single match state and D2 represents a gap after all match states have occurred. State X represents a gap internal to S(6, 1′), meaning a state M should occur on both sides of any state X.