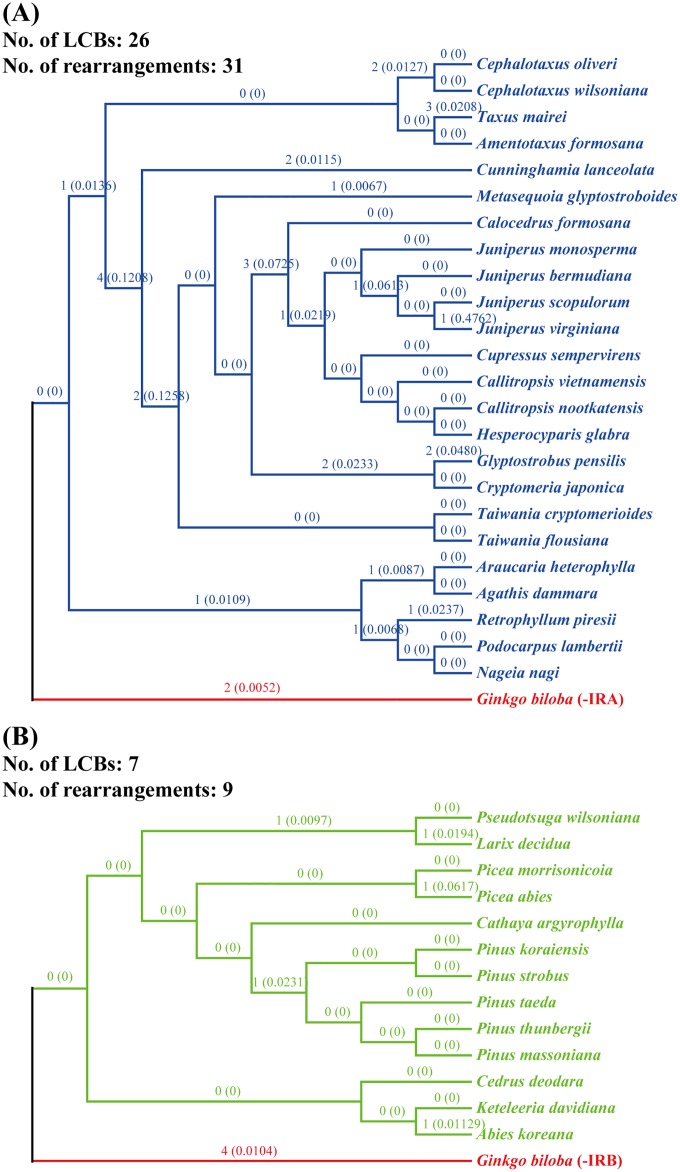
Fig 2. Chloroplast genome rearrangement estimates among cupressophytes and Pinaceae.
The topologies of clades cupressophytes (A) and Pinaceae (B) constructed from phylogenetic analysis were used as suggested actual trees and rearrangements were inferred from the matrices of cp genome LCBs. The estimated number of rearrangements for branches to taxa are shown above branches and corresponding rearrangements per million years were shown in brackets. The cp genome of G. biloba with removing IRA and IRB separately was used as a rooted genome in these two comparisons, respectively.