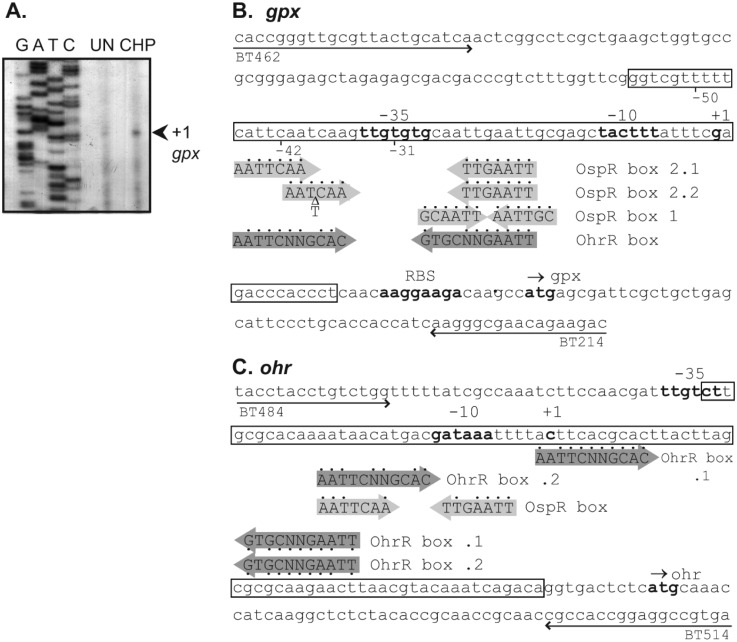
Fig 4. ospR and ohr promoter analysis.
(A) Primer extension analyses of gpx using total RNA extracted from Pseudomonas aeruginosa without and with 200 μM cumene hydroperoxide induction. The arrow indicates the transcription start site. The Sanger sequencing ladder to the left of the primer extension lanes was generated using the same primer used in the primer extension. The nucleotide sequences of the gpx (B) and ohr (C) promoter regions are shown. The putative −10 and −35 promoter regions, the transcription start site (+1), and the putative translation start codon are shown in bold. Locations, directions and names of oligonucleotides used in this study, i.e. BT462, BT214, BT484, and BT514, are indicated. The DNaseI-protected regions for OspR binding are boxed. Putative OspR binding sites and OhrR binding site are aligned with the predicted conserved sequences in shaded arrows. Dots indicate matched nucleotides.