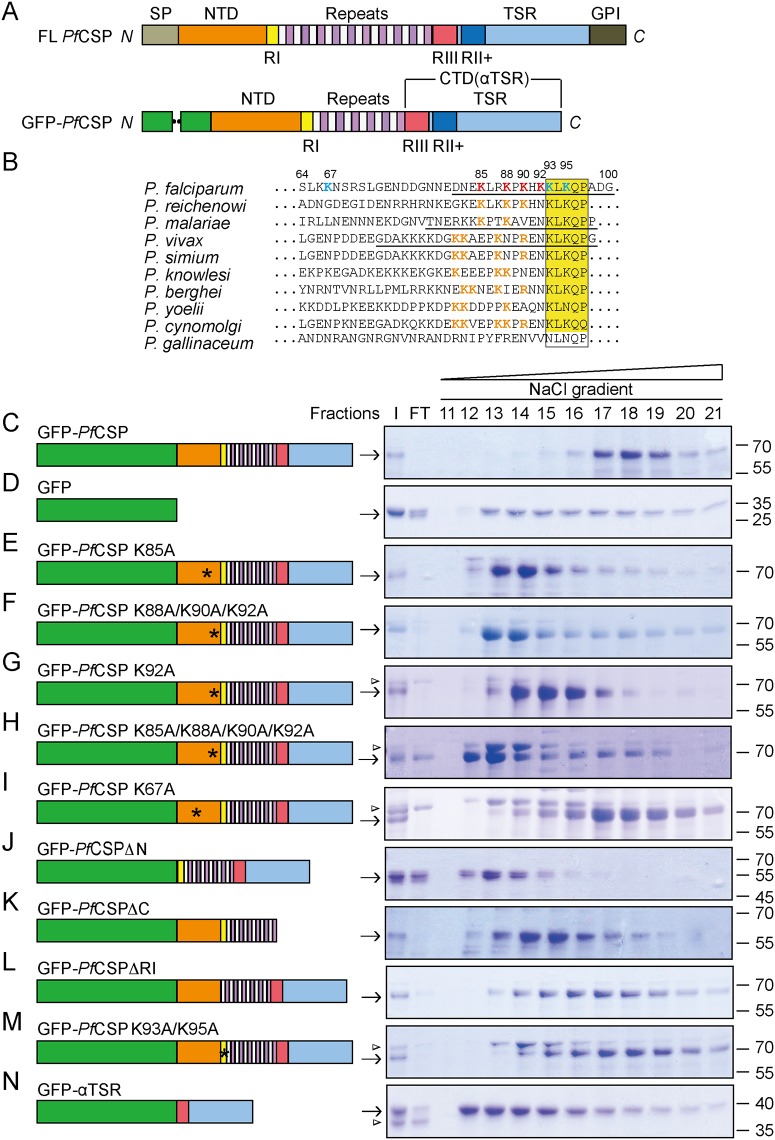
Fig 1. GFP-PfCSP interacts with heparin.
A, Domain structures of full-length and recombinant PfCSP. The termini are indicated. SP, signal peptide; NTD, N-terminal domain; CTD, C-terminal domain; RI, region I; RII+, region II plus; RIII, region III; GPI, GPI anchor sequence. B, Sequence alignment of residues preceding the region I in the NTD of CSP. Residues in PfCSP are numbered, and residues mutated are highlighted in bold. Mutations that affect heparin binding are colored in red and mutation that does not is colored in cyan. Basic residues in other CSPs that are near region I are colored in orange. The region I is highlighted by a yellow box. Peptides that have been tested for heparin binding are underlined. C,D, Heparin binding of GFP-PfCSP (C) and GFP alone (D). ~150 μg of purified protein was applied to the heparin column, and samples were analyzed by SDS-PAGE, followed by Coomassie staining. Domain structure of the protein used is shown on the left. I, input; FT, flow-through. E-N, as in C, but with GFP-tagged CSP mutants. Arrowhead indicates a contaminant. All data were confirmed by at least three independent experiments using three independently purified batches of proteins. Data shown are from a representative experiment.