Abstract
In this study, we analyzed the expression profile of four genes (CCNA2, CCNB1, CCNB2, and CDK1) in laryngeal squamous cell carcinoma (LSCC) cell lines and tumor samples. With the application of microarray platform, we have shown the overexpression of these genes in all analyzed LSCC samples in comparison to non-cancer controls from head and neck region. We have selected CDK1 for further analysis, due to its leading role in cell cycle regulation. It is a member of the Ser/Thr protein kinase family of proven oncogenic properties. The results obtained for CDK1 were further confirmed with the application of reverse transcription quantitative polymerase chain reaction (RT-qPCR) technique, Western blot, and immunohistochemistry (IHC). The observed upregulation of CDK1 in laryngeal squamous cell carcinoma has encouraged us to analyze for genetic mechanisms that can be responsible this phenomenon. Therefore, with the application of array-CGH, sequencing analysis and two methods for epigenetic regulation analysis (DNA methylation and miRNA expression), we tried to identify such potential mechanisms. Our attempts to identify the molecular mechanisms responsible for observed changes failed as we did not observe significant alterations neither in the DNA sequence nor in the gene copy number that could underline CDK1 upregulation. Similarly, the pyrosequencing and miRNA expression analyses did not reveal any differences in methylation level and miRNA expression, respectively; thus, these mechanisms probably do not contribute to elevation of CDK1 expression in LSCC. However, our results suggest that alteration of CDK1 expression on both mRNA and protein level probably appears on the very early step of carcinogenesis.
Electronic supplementary material
The online version of this article (doi:10.1007/s13277-016-4991-4) contains supplementary material, which is available to authorized users.
Keywords: CDK1, Overexpression, Gene methylation, microRNA, Western blot, Immunohistochemistry
Introduction
Larynx squamous cell carcinoma (LSCC) is one of the most frequent types of head and neck cancer [1]. The main etiological causes of this type of cancer are well known and include the synergistic effects of tobacco smoking and alcohol abuse [2].
Chromosomal aberrations resulting in alterations in gene expression (downregulation of tumor suppressor genes or overexpression of oncogenes) are one of the genetic factors contributing to the development of this cancer. Until now, several oncogenes have been shown to be involved in the development of head and neck cancers including CCND1, FGF3, FGF4, CTTN [3], or EGFR [4]. In our previous work, we have shown also other oncogenes like the ORAOV1, FADD [5], and CRKL [6] as potentially related to LSCC.
As aberrant cell cycle control plays a significant role in tumor development, we focused on the genes involved in this proces. We analyzed the expression level of group of four genes (CDK1, CCNB1, CCNB2, and CCNA2) engaged in cell cycle control. CDK1 (cyclin-dependent kinase 1) gene is an important factor of the cell cycle control system [7, 8]. Together with cyclin B (CCNB1 and CCNB2), it forms an active MPF (maturation-promoting factor) which makes cell enter mitosis [9]. The A cyclin (CCNA2) binds CDK1 during the transition from G2 to M phase [10]. Moreover, CDK1 is required for mammalian cell proliferation as it is the only CDK that can initiate the onset of mitosis [11]. For all these genes, we observed gene upregulation in LSCC cell lines and tumor samples as compared to non-cancer controls. However, due to the leading role of CDK1 in cell cycle control and regulation, we have chosen the CDK1 for further analysis. This gene is a member of the Ser/Thr protein kinases family. The oncogenic potential of these enzymes was demonstrated [12–14]. The CDK1 protein is a catalytic subunit of M-phase promoting factor essential for the G1/S and G2/M transition in eukaryotic cell cycle.
Interesingly, in model organism Saccharomyces cerevisiae, the 75 target genes of Cdk1 were shown to control cell cycle, DNA replication and segregation, transcriptional programs, and cell morphogenesis [15]. These findings suggest that deregulation in CDK1 gene may play a significant role in cancerogenesis. Moreover, the involvement of CDK1 in tumorgenesis was postulated in various types of cancer, including laryngeal cancer [16, 17].
Due to the suggested role of CDK1 in tumorigenesis, we analyze here the CDK1 DNA copy number, gene sequence, DNA methylation status, and miRNA expression profile with the aim to identify the responsible mechanism for the observed CDK1 upregulation in LSCC.
Materials and methods
Cell lines
Twenty-five cell lines derived from laryngeal cancer were used. The cell line characteristics are shown in Table 1.
Table 1.
Cell line characteristics
| Cell line number | Sex | Age (years) | Primary tumor location | TNM | Specimen site | Type of lesion | Grade | Survival |
|---|---|---|---|---|---|---|---|---|
| UT-SCC-6A | F | 51 | Supraglottic larynx | T2N1M0 | Larynx | rec | G1 | DWD 31 months |
| UT-SCC-6B | F | 51 | Supraglottic larynx | T2N1M0 | Neck | met | G1 | DWD 31 months |
| UT-SCC-8 | M | 42 | Supraglottic larynx | T2N0M0 | Larynx | pri | G1 | DWD 35 months |
| UT-SCC-11 | M | 58 | Glottic larynx | T1N0M0 | Larynx | rec | G2 | DNE >5 years |
| UT-SCC-13 | M | 53 | Supraglottic larynx | T3N0M0 | Larynx | rec | G2 | DWD 11 months |
| UT-SCC-19A | M | 44 | Glottic larynx | T4N0M0 | Larynx | pri | G2 | DNE >5 years |
| UT-SCC-19B | M | 44 | Glottic larynx | T4N0M0 | Larynx | pri (per) | G2 | DNE >5 years |
| UT-SCC-22 | M | 79 | Glottic larynx | T1N0M0 | Larynx | rec | G2 | DWD 28 months |
| UT-SCC-23 | M | 66 | SCC transglottic | T3N0M0 | Larynx | pri (per) | G1 | DNE >5 years |
| UT-SCC-29 | M | 82 | Glottic larynx | T2N0M0 | Larynx | pri | G1 | DNE 10 years and 4 months |
| UT-SCC-34 | M | 63 | Supraglottic larynx | T4N0M0 | Supraglottic larynx | pri | G1 | DWD 10 months |
| UT-SCC-35 | M | 50 | Glottic larynx | T2N0M0 | Larynx | resid | G2 | DWD 10 months |
| UT-SCC-38 | M | 66 | Glottic larynx | T2N0M0 | Larynx | pri | G2 | DWD 16 months |
| UT-SCC-42B | M | 43 | Supraglottic larynx | T4N3M0 | Neck | pri | G3 | DWD 2 months |
| UT-SCC-49 | M | 76 | Glottic larynx | T2N0M0 | Larynx | pri | G2 | DWD 2 years and 7 months |
| UT-SCC-50 | M | 70 | Glottic larynx | T2N0;rT2N0 | Larynx | rec | G3 | ANE >5 years |
| UT-SCC-57 | M | 76 | Glottic larynx | T2N0M0 | Larynx | rec | G1-G2 | DWD 4 years |
| UT-SCC-75 | M | 56 | SCC laryngis | T2N2BM0 | Larynx | pri | G2 | D, NED 2 years and 6 months |
| UT-SCC-106A | M | 59 | SCC plicae vocalis | T1AN0M0 | Larynx | pri | G2 | DNE second pri 4 years and 1 month |
| UT-SCC-106B | M | 59 | SCC plicae vocalis | rT3N0M0 | Larynx | rec | G3 | DWD 5 days |
| UT-SCC-107 | M | 46 | SCC laryngis supraglottis | T4N2CM0 | larynx | pri | G2 | DNE 19 months |
| UT-SCC-108 | M | 68 | SCC laryngis supraglottis | T2N0M0 | larynx | pri | G3 | DWD 19 months |
| UT-SCC-113 | M | 50 | SCC laryngis transglottica | T3N0M0 | larynx | pri | G3 | DWD 17 months |
| UT-SCC-116 | M | 60 | SCC laryngis supraglottis | T4N1M0 | larynx | pri | G2 | DWD 9 months |
| UT-SCC-117 | M | 71 | SCC laryngis (resid T2N0M0) | T2N0M0 | Larynx | rec | G2 | DWD 47 months |
All cell lines were obtained in University of Turku (Finland)
M male, F female, TNM TNM classification (T tumor, N lymph node involvement, M distance metastases), pri primary tumor, rec recurrence, met metastasis, per persistent tumor, DWD died with the disease, DNE died with no disease evident, ANE alive with no disease evident
Primary tumor samples
Primary tumor samples used for the mRNA and microRNA expression analysis and pyrosequencing
Forty-five laryngeal cancer samples (1 female and 44 males) were used in the study. The average patients age was 61 years (ranged 42–84). The TNM and G status details are shown in Table S1. During the surgery, each sample was divided into three parts and designated for histopathological analysis, DNA isolation (immediate freezing in −80 °C), and RNA analysis (storage in RNAlater, Sigma, according to manufacturer’s instruction). Only samples containing more than 60 % of tumor cells were chosen for the study. The study was approved by the local ethical board of Medical University in Poznan. Written consent was obtained from all donors.
Primary tumor samples used for immunohistochemistry
The studies were performed on a group of 40 patients (5 females, age, 50–69 and 35 males, age, 44–77), who underwent total laryngectomy. Based on histopathological examination, performed by two independent pathologists, in all cases, laryngeal squamous cell carcinoma (LSCC) was diagnosed. The tumor stage was determined according to the current TNM classification published by the International Union Against Cancer (IUAC). The TNM and G status details are shown in Table S1.
The immunohistochemical studies were performed on selected archival formalin-fixed paraffin embedded (FFPE) tissue sections. All cases were revised and selected by two independent pathologists according to hematoxylin and eosin-stained tissue sections. In each sample, cancer cells occupied approximately 80 % of tissue area.
Control samples
Control samples used for the expression analysis
Various types of non-malignant samples were used as controls for microarray and reverse transcription quantitative polymerase chain reaction (RT-qPCR) expression analysis: the commercially available human total RNA derived from healthy larynx (Total Larynx RNA, Stratagene, Agilent Technologies, Waldbronn, Germany), the RNA derived from bronchial airway epithelia reconstituted in vitro (two donors) (EC, Epithelix Sarl, Geneve, Switzerland), normal mucosa derived from surgical margin during laryngectomy (LX10), normal human bronchial/tracheal epithelial cells—NHBE (Lonza, Verviers, Belgium), and human tracheal epithelial cells—HTEC (PromoCell, Heidelberg, Germany).
Control samples used for CDK1 gene promoter DNA methylation analysis
Two groups of controls included 20 DNA samples isolated from head and neck region. Ten were derived from the oral cavity epithelium (buccal swabs, W1–W10) from healthy donors. The second group of controls (K1–K10) were collected during surgeries not associated with cancer and include samples from the resected epithelium of Reinke’s edema and normal vocal fold fragments removed during surgical widening of the glottis in patients with bilateral vocal cord paralysis. Additionally, the fully methylated standard (MK, Millipore, Hilden, Germany) and unmethylated DNA (UM), i.e., the whole genome amplified DNA from peripheral blood lymphocytes (prepared with GenomePlex® Whole Genome Amplification Kit) were used in each run.
Protein lysates used for Western blot analysis
The presence of the examined protein (CDK1) was analyzed in comparison to commercially available total larynx tissue lysates (Larynx Human Tissue Lysates from adult normal tissue—ab44731 and ab44733, pooled, Abcam, UK), four protein lysates obtained from the non-cancer tissue samples (K12-K15), and three lysates from the epithelial cells (primary culture obtained in our laboratory) derived from the non-cancer tissue samples (K2, K3, K6). As positive controls, the Jurkat cell line lysate (Jurkat E6.1, Human T-cell lymphocytes, ECACC, Salisbury, UK) or HeLa lysate (cervical cancer cell line) was used.
Control samples used for the immunohistochemical analysis of CDK1 protein
The control material for the immunohistochemical studies consisted of archival FFPE tissue sections, revised, and selected by two independent pathologists. The sections contained a disease-free normal mucosa, at least 2 cm distant from the tumor margins. To establish the immunohistochemical protocol, a series of positive and negative control reactions were performed. The positive control reaction was performed on placenta, where the presence of CDK1 protein expression was indicated (reference sources: [18]; the Human Protein Atlas; and manufacturer antibodies datasheet). The negative control reaction was performed by substituting the primary antibody by the 1 % BSA (bovine serum albumine) solution in PBS (phosphate buffered saline).
For all the control samples used in our study, an appropriate agreement from the bioethical board of the Poznan Medical University was obtained.
Cell culture
The culture condition for cell lines and primary cultures of epithelial cells were described elsewhere with minor modification [5, 19, 20]. For DNA, RNA, and protein isolation, cell lines were cultured to 80 % confluence and then harvested with 0.1 % trypsin and 0.2 % EDTA. The cell lines used as controls for Western blot were cultured under identical conditions, using the modified Eagle medium (MEM) for HeLa and RPMI-1640 medium for the Jurkat cell line, with 100 U/mL penicillin/streptomycin medium, 37 °C, 5 % CO2).
DNA, RNA, and protein isolation
Nucleic acids from cell lines, tumor samples, and peripheral blood lymphocytes were isolated according to standard methods: phenol/chloroform extraction and ethanol precipitation for DNA and Chomczynski’s method with application of Trisol for RNA [21]. Nucleid acids purity and concentration was analyzed as described elsewhere [22].
Total protein extraction from the LSCC cell lines and controls (except tissue samples) was performed with Nonidet-P40 (NP-40) buffer with the Protease Inhibitor Cocktail (LabEmpire, Rzeszow, Poland). Total protein lysates from the non-cancer tissue samples were obtained with the use of FASTPREP-24™ 5G Instrument (MP Biomedicals) after the grinding in liquid nitrogen Protein concentration was measured with standard Bradford method [23].
Microarray-based gene expression analysis
Expression analysis was performed on GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix) for 10 cell lines (UT-SCC-6A, −11, −19B, −22, −29, −34, −57, −106A, −107, −116), 5 tumor samples (out of 45 collected), and 3 control samples (Total Larynx RNA, EC, LX10). The microarrays were performed and analyzed as described elsewhere [5, 24, 25]. The number of tags used to analyze the expression of chosen genes was as follows: two tags for the CCNA2 gene (203418_at and 213226_at), one for CCNB1 (214710_s_at), and one for CCNB2 (202705_at). Two tags were used to analyze CDK1 expression: 203213_at and 203214_x_at as they correspond to entire coding exons of the gene. The gene upregulation was defined as increased gene expression observed for analyzed tag in comparison to expression level of this tag in at least two of the three non-cancer controls.
Reverse transcription and quantitative real-time PCR
Total RNA from 25 LSCC cell lines and 3 non-tumor control samples (Total Larynx RNA, NHBE cells and HTEC cells) were used to synthesize the cDNA template using of Enhanced Avian RT First Strand Synthesis Kit (Sigma-Aldrich), according to manufacturer’s protocol. RT-qPCR primers were designed with the use of Beacon Designer™ 7.5 (PRIMER Biosoft International) and the specificity was verified with the primer BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Three genes were used as reference genes: ARNT, UBC, and GAPDH. The primer sequences are listed in Table S2. Quantitative real-time PCR was performed with the iCycler iQ5 (Bio-Rad), and the gene expression profile was analyzed with a detection system using iQ5 Optical System Software 2.0 (Bio-Rad Laboratories). The annealing temperature applied for CDK1 and reference genes was 55 °C; detailed PCR conditions are presented in Supplementary data. For PCR, the 5× HOT FIREPol® EvaGreen® qPCR Supermix (Solis BioDyne, Estonia) according to the manufacturer’s protocol was used. For each sample, 0.4 μL of cDNA was used (undiluted reverse transcription product derived from 8 μg RNA in 40 μl reaction). The melting curve, determination of PCR efficiency, PCR data analysis, and statistics were previously described [6]. To estimate the changes in gene expression, the cutoff point value was calculated according to the scheme: three-times standard deviation of the controls plus the expression value of the highest control.
CDK1 protein western blot analysis
The whole cell lysates from 25 cell lines and 8 samples from the non-cancer larynx tissue were used. In each blot, positive control (Jurkat and/or HeLa cell line lysate) was used. Detailed information can be found in the Supplementary data. The rabbit polyclonal anti-CDK1-C-terminal antibody (ab7953, Abcam, UK, dilution 1:1000) was applied for all 25 cell lines and 8 non-cancer larynx tissue lysates. The second, N-terminal antibody (ab131011, Abcam, UK, dilution 1:6000) was used for the analysis of 7 LSCC cell lines (of the 25 collected) and total larynx tissue lysate. Furthermore, the anti-CDK1 (phospho T161) antibody (ab138389, Abcam, UK, dilution 1:750) was applied for 24 LSCC cell lines. Blots were incubated overnight in 4 °C and secondary antibody (goat anti-Rabbit, ab 97051, Abcam, UK, 1:38500) was applied (2 h, room temperature). Rabbit anti-GAPDH (ab 9485, Abcam, UK, dilution 1:2500) antibody was used as a loading control. For the protein detection, SuperSignal West Pico Chemiluminescent Substrate (Thermo Scietific, Rockford, IL USA) was used and the images were scanned and analyzed with the ChemiDoc XRS+ System (BioRad).
Immunohistochemical analysis of laryngeal cancer formalin-fixed paraffin embedded tissue sections
The level of CDK1 protein expression was performed using automated morphometric methods in ImageJ 1.46a Program, using authors’ macro described earlier [26]. The representative microphotographs were taken at ×20 original objective magnification in the light microscope ECLIPSE E800 (Nikon Instruments Europe, Amsterdam, Netherlands), with Nikon Digital Sight DS-5Mc camera (Nikon Instruments Europe, Germany) driven by 4NIS Elements F 3.0 software (Nikon Instruments Europe, Germany). First, taken microphotographs were converted using “Colour Deconvolution” option, where the brown pigment was “isolated” from microphotography. Subsequently, using the “Threshold” option, the microphotography was converted into an 8-bit version, which allowed to creation of so-called mask. The created “mask” was applied on the original microphotography, and conversions of the color intensity into numerical data were performed. The level of CDK1 protein expression was evaluated according to modified immunoreactive scale (IRS) described first by Remmele and Stegner [27] and used (partially in modified version) in our previous publications [28–30]. The IRS was evaluated as the ratio of the percentage of positive stained cells/area (PP) and the intensity of the color reaction (SI) (IRS = SI × PP).
Microarray DNA copy number analysis (array-CGH)
Array-CGH profiles from 13 cell lines from our previous study were used [5, 24, 25].
The log2ratio for the DNA region: chr10:62,208,242-62,223,930 covering the CDK1 gene (according to NCBI36/hg18) Assembly) was analyzed. The mean log2 ratio of tags for the DNA region containing the CDK1 gene was calculated. The mean log2ratio value between 0.5 and −0.5 was assumed as normal.
The analysis of CDK1 gene promoter DNA methylation by bisulfate pyrosequencing
DNA samples from 25 cell lines, 41 tumor samples, and controls (oral epithelium—buccal swabs, normal head, and neck tissue samples) were used. Purified DNAs were converted with bisulfite solution with the use of EpiTect DNA Modification Kit (QIAGEN, Germany), according to the manufacturer’s protocol. The primer designing and pyrosequencing protocol was performed as described elsewhere [22]. Fully methylated and unmethylated controls were used in each run. Seven CpG sequences in one CpG island were analyzed. See Supplementary data for PCR details and cutoff points calculation method. Samples with DNA methylation level above the established upper cutoff point and below the lower cutoff point were regarded as hyper- and hypomethylated, respectively.
Sequencing analysis
Primer pairs for mutation analysis of coding exons and intron–exon junctions of CDK1 gene were designed with the use of Primer3 v.0.4.0 online tool (http://bioinfo.ut.ee/primer3-0.4.0/). Primer details are listed in Table S3. Detailed PCR conditions are available in Supplementary data. PCR products were sequenced using Big Dye Terminator Sequencing Kit Cycle v3.1 (Applied Biosystems, Inc. (ABI), Foster City, CA, USA), according to the manufacturer’s protocol and separated using ABI PRISM 310 Genetic Analyzer (Applied Biosystems). The results were analyzed using Sequencing Analysis v. 5.2-5.4 software’s and the CodonCode Aligner software (demo mode). The reference sequence for CDK1 gene was obtained from RefSeq database; NM_001786; UCSC Genome Browser GRCh37/hg19.
Microarray-based microRNA expression analysis
MicroRNA expression analysis was performed using the Agilent Human microRNA Expression Microarray 66K (based on miRBase 16.0 and updates) on 16 LSCC cell lines, 5 representative primary LSCC tumor samples (out of 45 collected), and 3 non-tumor controls (Total Larynx RNA, NHBE cells and HTEC cells). To delineate the microRNAs with the highest probability to regulate CDK1 gene, we used the following selection criteria: (1) significantly up or downregulated miRNAs, with at least twofold change as compared to controls, delineated by miRNA expression microarrays, (2) miRNAs predicted in silico to target CDK1 by at least three miRNA databases, using MiRWalk tool (from http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/index.html) and showing at least eight seed nucleotide homology (p < 0.05).
Results
Microarray-based gene expression analysis
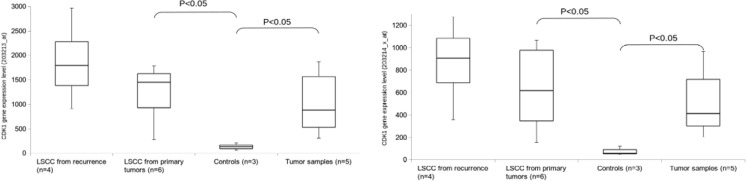
For all chosen genes, the expression level was higher in both cell lines and tumor samples as compared to non-cancer controls. For analyzed cyclins, the gene upregulation was shown in 9/10 (90 %) LSCC cell lines and 5/5 (100 %) tumor samples (data not shown). Among them, we choose CDK1 for further analysis, due to its leading role in cell cycle regulation and proven oncogenic potential of this group of enzymes. For the CDK1 gene, the analyzed tags (203213_at and 203214_x_) indicated higher expression of the CDK1 gene in all analyzed cell lines, tumor samples, and controls. The expression level was higher both in cell lines and in tumor samples as compared to non-cancer controls (p < 0.05; Mann-Whitney U test, Fig. 1). No statistically significant difference between cell lines derived from primary and recurrence tumors was observed, but the CDK1 gene expression level was higher in primary tumor cell lines as compared to controls (p < 0.05, U test for both tags, Fig. 1). Thus, significantly higher expression level was observed in tumor samples and cell lines compared to non-cancer controls.
Fig. 1.
The CDK1 gene expression analysis (microarray). Box plots show gene expression level in cell lines derived from primary tumors, recurrent tumors, and tumor samples in reference to non-cancer controls from head and neck region. Figure presents results obtained for two most representative tags: 203213_at (left) and 203214_x_at (right). The Mann–Whitney U test was performed
Quantitative real-time PCR
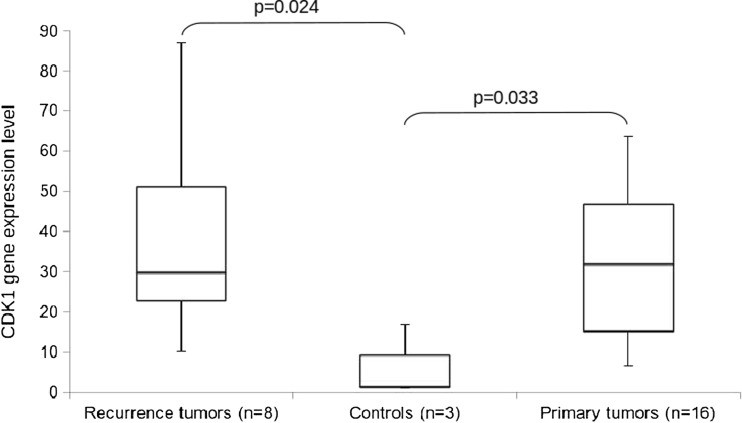
To confirm the overexpression of the CDK1 gene, the quantitative real-time PCR technique was performed on LSCC cell lines and non-cancer controls from head and neck region. Higher expression levels of the CDK1 gene has been demonstrated in 9/25 (36 %) cell lines as compared to three non-cancer controls (p < 0.05; Mann-Whitney U test, Fig. 2).
Fig. 2.
The box plot of RT-qPCR results showing CDK1 gene expression levels in cell lines derived from laryngeal cancer recurrence tumors, primary tumors, and controls. The Mann–Whitney U test was performed. UT-SCC-6B cell line was excluded from this part of analysis as it is derived from metastasis tumor
Statistically significant difference between expression in LSCC cell lines in comparison to controls was observed, but there is no significant difference between cell lines derived from primary tumors and recurrent tumors (Fig. 2). The results confirmed our initial observation of the CDK1 gene overexpression in LSCC. No correlation of CDK1 gene expression level with tumor phenotype (TMN and grade status) or patients survival was observed.
Western blot analysis results
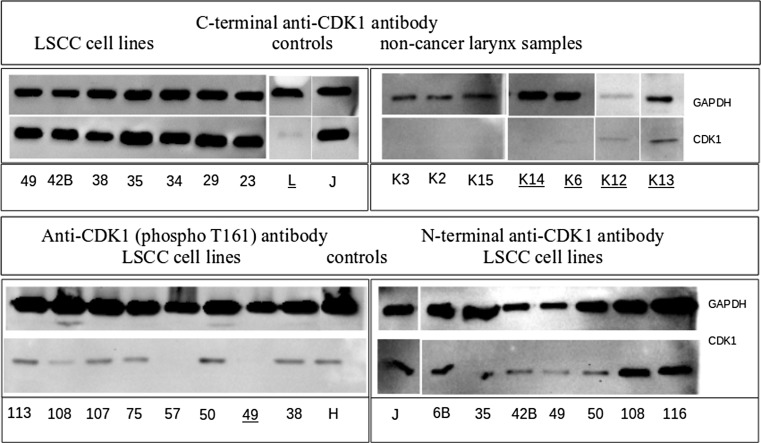
To determine the expression of CDK1 on protein level, Western blot analysis was performed. As was shown with the C-terminal anti-CDK1 antibody, the CDK1 protein was present in all 25 analyzed cell lines. Trace amount of CDK1 protein was detected in five of the non-cancer larynx tissue samples (i.e., K6, K12, K13, and K14 and total larynx tissue lysate.) The N-terminal anti-CDK1 antibody has shown the presence of CDK1 protein in all seven LSCC cell line lysates but absent in the total larynx lysate. The analysis of 24 LSCC cell lines with the application of anti-CDK1 (phospho T161) antibody has demonstrated the presence of phosphorylated CDK1 protein in 22 LSCC cell lines; in 1 LSCC cell line (UT-SCC-49) trace amount of analyzed protein was observed; and in one LSCC cell line (UT-SCC-57), lack of phosphorylated CDK1 protein was shown. The positive control lysates has demonstrated the presence of CDK1 protein, and the loading control—GAPDH protein was present in all analyzed tissue lysates. For all the analyzed proteins, one band with adequate size was observed (Fig. 3).
Fig. 3.
Western blot results obtained for different types of anti-CDK1 antibodies. The anti-CDK1 C-terminal antibody (upper panel) was applied for analysis of LSCC cell lines (left) and non-cancer head and neck tissue (right) lysates, the anti-CDK1 (phospho T161) antibody (lower panel, left) and the N-terminal anti-CDK1 antibody (lower panel, right) were applied on the LSCC cell line lysates. The numbers used are the LSCC numbers; K2, K3, K6, K12–K15 tissue sample donors numbers, L total larynx tissue lysate, J Jurkat cell line lysate, H HeLa cell line lysate. GAPDH was used as a loading control and Jurkat and HeLa cell lines were the positive controls. The samples where the trace amounts of CDK1 protein were observed are underlined
This analysis has shown the presence of the CDK1 protein in LSCC and its absence or small amounts in controls derived from non-cancer tissue from the larynx or regions surrounding the larynx. As was shown, in LSCC, the CDK1 is present in active-phosphorylated form. Moreover, this analysis shown the lack of the changes in protein length resulting from the CDK1 gene polymorphism rs3212319.
IHC analysis results
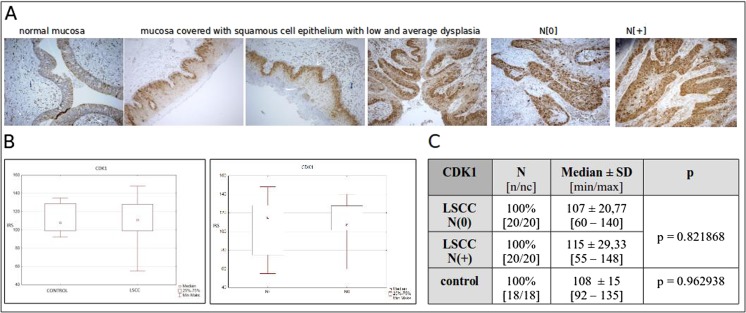
The immunohistochemical studies were performed to evaluate the localization of CDK1 protein expression in LSCC primary tumors, as well as for description of the differences in protein expression (cytoplasmic vs. nuclear) in research LSCC group of patients. The analyses showed nuclear-cytoplasmic expression of CDK1 protein in all 40/40 (100 %) primary LSCC and in all 18/18 (100 %) cases of normal mucosa (Fig. 4a), but higher CDK1 expression in LSCC tumors median [CDK1 IRS = 111.00] compared to normal mucosa median [CDK1 IRS = 108.00] was observed. In 20/40 LSCC cases, we have revealed the nuclear expression of CDK1 protein; the remaining 20/40 cases revealed only cytoplasmic expression of analyzed antigen. No significant differences in expression of the studied protein was observed between primary LSCC and control group (p = 0.962938) (Fig. 4b,c).
Fig. 4.
Immunohistochemical representative microphotographs representing the cdk-1 expression in normal mucosa (represented as columnar epithelium); on the following three micrographs, there are presented fragments of mucosa covered with squamous cell epithelium with low and average dysplasia, then primary LSCC in patients without lymph node metastases N (0), in LSCC in patients with lymph node metastases N (+). Brown color corresponds to localization of CDK1 antigen, antibody complex visualized by peroxidase system, and developed by DAB as a chromogen, nucleus counterstained with hematoxylin. Primary objective magnification ×10
According to lymph node involvement, we have revealed lower CDK1 protein expression in patients without lymph node metastases (CDK1IRS N (0) = 107) compared to the patients with lymph node involvement (CDK1IRS N (+) = 115) (Fig.4b,c). Moreover, in LSCC N (0) group, 9/20 cases revealed nuclear expression of CDK1, and in LSCC N (+) group, 11/20 cases revealed the nuclear expression of established antigen. No significant differences in expression of the studied protein was observed in the analyzed groups (p = 0.821868). Whereas, no significant differences between nuclear and cytoplasmic expression, as well as lack of correlation between nuclear-cytoplasmic ratio of CDK1 expression according lymph node metastases was shown. The differentiated pattern of CDK1 expression profile may indicate the heterogeneity of the LSCC.
Microarray-based DNA copy number analysis
We first analyzed available array-CGH profiles to check for possible copy number gains of the CDK1 locus in the 13 LSCC cell lines. In all instances, the mean log2 ratio for analyzed LSCC cell lines ranged from −0.20 to 0.15. Therefore, in all 13 cell lines, DNA copy number changes in the CDK1 locus cannot account for the observed evaluated expression level of the CDK1 gene in LSCC.
CDK1 gene sequencing results
As no copy number alterations, that could explain elevated expression of CDK1, were identified, we screened for potential activating mutations in coding sequences of the gene. Two common SNP variants were found in the analyzed samples: rs3212319 (15/25 cell lines) and rs1871446 (24/25 cell lines). The rs3212319 polymorphism is an intronic variant (NC_000010.10: g.62551816delC; NM_001786.4: c.653 + 5delC; (GRCh37/hg19)) and results in a deletion of the cytosine in the position +5 in intron 6. Of the 15 affected samples, 13 were heterozygotes (C/-) and 2 were deleted in a homozygous manner (−/−). The rs1871446 polymorphism is a 3′UTR variant and results in a A > G substitution (NC_000010.10: g.62553763A > G; NM_001786.4: c.*30A > G; UCSC (GRCh37/hg19). Of the 24 affected samples, 4 were heterozygous alterations (A/G), while the rest were homozygotes (G/G). No other alterations in coding sequence or exon-intron junction site were detected. Therefore, regarding the expression data, most probably none of these SNP is associated with the elevated CDK1 observed in LSCC.
DNA methylation analysis of CDK1 promoter region by pyrosequencing
The changes in pattern of DNA methylation is one of the mechanisms of the gene transcription regulation. DNA methylation level of CDK1 gene promoter was analyzed in all cell lines and primary tumor samples. The buccal swabs and non-cancer head and neck tissue samples were used as controls. The normal DNA methylation level was estimated to range between 1.59 and 13.7 %. The mean methylation for the LSCC cell lines ranged between 3.33 and 6.81 % and for the tumor samples ranged 0 %–1.46 %. Therefore, no meaningful differences were found between the samples. Thus, the CDK1 gene hypomethylation was excluded as a mechanism of the gene upregulation.
Microarray-based microRNA expression analysis
Lastly, we analyzed another epigenetic mechanism that on account for changes in gene expression—miRNAs. We delineated the group of microRNAs with significant downregulated expression between 16 cell lines, 5 primary tumor samples, and 3 controls. The analysis has revealed 12 downregulated miRNA genes. None of this was reported to be involved in CDK1 gene regulation. Therefore, we failed to delineate any miRNA which could regulate CDK1 expression level in the analyzed cases.
Discussion
In the current study, we have analyzed the expression pattern of group of genes —CCNB1, CCNB2, CCNA2, and CDK1 in laryngeal squamous cell carcinoma. All of the chosen genes are important factors in eukaryotic cell cycle. We have shown that all the analyzed genes are upregulated in LSCC cell lines and tumor samples as compared to non-cancer head and neck controls. From this group of genes, we have chosen the CDK1 gene for further analysis in context of searching the new potential oncogenes in LSCC. The main criterion we assumed for choosing this gene was its role in cell cycle control. CDK1 is a member of the Ser/Thr protein kinase family, and its product is a catalytic subunit of M-phase promoting factor, essential for G1/S and G2/M phase transitions of eukaryotic cell cycle. The oncogenic character of Ser/Thr protein kinases like B-Raf and PIM1 was shown [12–14]. It was demonstrated that CDK1 is suifficient to drive the cell cycle in mammalian cells [31] which indicates the leading role of this kinase in cell cycle regulation. Moreover, as was shown in previous papers, CDK1 gene phosphorylates a group of transcription factors (TF) involved in cell cycle control and cell proliferation [32, 33].
Nowadays, large field of cancer studies focuses on the identification of novel, potential oncogenes or tumor suppressor genes. The detailed knowledge of tumor genetic background is fundamental for establishing new diagnostic and treatment strategies, including targeted therapy. Genetic data have been successfully transformed into clinical practice, for example for chronic myeloid leukemia (CML) where the application of BCR-ABL tyrosine kinase inhibitor—imatinib is widely used [34], and in case of HER2-positive breast cancer, combination of lapatynib and transtuzumab is applied for inhibition of HER2 [35]. So far, for head and neck tumors, only Cetuximab has been approved for treatment [36]. Here, we have analyzed a potential target for an inhibition therapy—CDK1 in larynx squamous cell carcinoma (LSCC) and show its overexpression in this tumor. Importantly, Goga et al. showed that the inhibtion of CDK1 gene leads to downregulation of survivin expression and induction of MYC-dependent apoptosis [37]. Moreover, the CDK1 gene silencing with the application of RNAi was indicated as a novel potential tool for the therapy of malignant pleural mesothelioma [38], epithelial ovarian cancer [39], and breast cancer [40, 41]. The in-depth analysis of our microarray expression profiles showed higher CDK1 expression in both larynx cancer cell lines and primary tumor samples in comparison to normal controls. This observation was further confirmed with the application of quantitative real-time PCR technique. This is in line with previous results showed by Lian et al. in larynx cancer tissues compared to adjacent non-neoplastic tissues [17]. Besides, CDK1 overexpression in cancer was shown also for oral squamous cell carcinoma [42], breast cancer [43], epithelial ovarian cancer [39], and hepatocellular carcinoma [44].
To confirm overexpression of CDK1 on protein level, we performed the Western blot analysis. Our results indicated that the expression of CDK1 protein is absent or very weak in most of the normal larynx tissue as compared to LSCC cell lines. Moreover, in LSCC cell lines, the CDK1 protein was characteristic for phosphorylation at threonine 161, and it was shown that this is a feature of CDK1 activation [45]. However, the immunohistochemical analysis of laryngeal cancer tissue sections has revealed that the CDK1 protein is present both in cancer and non-cancer tissue samples (histopathologically normal surgical margins of laryngeal tumors). We suppose that these inconsistencies result from the differences of routinely used control samples. For the Western blot analysis, the control samples consisted of commercially offered total larynx tissue lysates and samples derived from the non-cancer disease interventions in the vicinity of the larynx, and as such, material is free from cancerogenic process. In samples derived from the other non-cancer samples, the trace amounts of CDK1 protein were observed. On the other hand, the control samples applied in immunohistochemistry (IHC) studies were derived from surgical margins of laryngeal tumors. The literature data indicate that such material, even if histopathologically normal, may contain genetic alterations appearing as the result of field cancerization process [46]. This is exemplified by the fact that second primary tumors frequently develop in patients after laryngectomy. Thus, the surgery margins contain cells that can transform into cancer. It is probable then that deregulation of CDK1 gene expression and the altered protein level occurs on very early step of carcinogenesis. The role of CDK1 protein overexpression in surgery margins of laryngeal cancer was noticed by Yang and coworkers [16] who had shown its relation with the local relapse occurrence. The CDK1 protein overexpression in OSCC was also shown in Xin Chen group, who had shown the presence of CDK1 protein in non-cancer epithelium in 35 % of OSCC cases and in 67.5 % of oral cancer samples [47].
We have also analyzed the nuclear-cytoplasmic ratio of CDK1 expression. In our study, no significant changes were found in the LSCC group as compared to normal mucosa samples. Also, no differences were shown within the LSCC samples group when the N0 group was compared to N+ group. This is in contrast to findings from colorectal tumor when it has been shown that high nuclear/cytoplasmic ratio of CDK1 expression is connected with poor prognosis in this type of cancer [48].
We further seek to identify mechanisms that potentially underlay the altered expression of CDK1 gene. We searched for activated mutation by Sanger sequencing that could possible explain CDK1 overexpression. In this analysis, two nucleotide polymorphisms (SNP)—rs1871446 and rs3212319 were detected. In our study, in 25 LSCC cell lines for polymorphism rs1871446, 1 homozygous (A/A), 4 heterozygous alterations (A/G), and 20 homozygotes (G/G) were detected. No correlation of analyzed SNP with patient survival time was observed; thus, we did not confirm any influence of GG homozygotes on the course of laryngeal cancer. Interestingly the second polymorphism, rs3212319 results in a deletion of cytosine in +5 position of intron 6. Of 15 affected samples, 13 were heterozygotes (C/−), and 2 were deleted in a homozygous manner (−/−). To verify whether this sequence variation induces changes in CDK1 protein length, the N-terminal anti-CDK1 antibody was used for the Western blot approached. Lysates of seven LSCC cell lines (two heterozygotes and two altered homozygotes) were used. The results of this analysis suggest that rs3212319 has no influence on CDK1 protein length. Moreover, no correlations of the observed polymorphisms with expression level of CDK1 gene on both mRNA and protein level in LSCC cell lines were found.
Therefore, using array-CGH, we excluded that gain of copy number of the CDK1 gene is responsible for the transcriptional upregulation. Further, we found no differences in the gene promoter methylation level between in LSCC cell lines, primary tumors, and control samples. Therefore, we conclude that lack of DNA methylation in CDK1 promoter region comprises a normal condition found in healthy controls and thus cannot be considered as the mechanism driving CDK1 overexpression.
Lastly, we hypothesized that CDK1 overexpression may be connected with downregulation of a microRNA that regulate the gene. It is already known that microRNA-7 and miR-490-3-p target CDK1 gene [49, 50]. Interestingly, microRNA-7 is downregulated in adrenocortical carcinoma and transfected to the cells and leads to the inhibition of CDK1 gene. Moreover, a group of other microRNAs was identified as potentially involved in the regulation of CDK1 gene in laryngeal cancer. Among them, the hsa-miR-139, hsa-miR-203, and miR-145 downregulation were observed in larynx and hypopharynx tumor samples [51, 52]. However, our study did not confirm these data—no changes in the expression profile of any of these microRNAs were observed using a microarray profiles.
Conclusion
In summary, our results show that CDK1 gene expression is recurrently elevated in LSCC. Moreover, it suggests that the alterations may appear in the very early step of carcinogenesis, as its mRNA and protein abundance is observed in surgery margin and T1 tumors. These data and the important role of CDK1 in cell cycle regulation and control as well as the proven impact of gene silencing on the tumor cell growth imply that this gene may function as a potential oncogene. However, the confirmation of the oncogenic character of CDK1 gene requires further study. Our attempts to identify the molecular mechanisms failed because we did not observe significant changes neither in the DNA sequence nor in the gene copy number. Similarly, the pyrosequencing and miRNA expression analyses did not reveal any differences in methylation level and miRNA expression, respectively; thus these mechanisms probably do not contribute to elevated of CDK1 expression observed in LSCC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 18 kb)
(DOCX 14 kb)
(DOCX 15 kb)
(DOCX 15 kb)
Acknowledgments
This work was funded by the National Science Centre, number N N403 153140, NN401195839 and Scholarship support for Ph.D. students specializing in majors strategic for Wielkopolska’s development for year 2011/2012, Sub-measure 8.2.2 Human Capital Operational Programme, co-financed by European Union under the European Social Fund
We thank prof. Reiner Siebert (Institute of Human Genetics, University Hospital Schleswig-Holstein Campus Kiel, Christian-Albrechts University 24105 Kiel, Germany) for making the microarray CGH experiments possible.
Compliance with ethical standards
Ethical standards
We declare that all experiments were performed in accordance with the current law of Poland. The study was approved by the local ethical board of Medical University in Poznan.
Conflicts of interest
None.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Maurya SS, Katiyar T, Dhawan A, Singh S, Jain SK, Pant MC, et al. Gene-environment interactions in determining differences in genetic susceptibility to cancer in subsites of the head and neck. Environ Mol Mutagen. 2015;56(3):313–21. doi: 10.1002/em.21920. [DOI] [PubMed] [Google Scholar]
- 3.Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 4.Rikimaru K, Tadokoro K, Yamamoto T, Enomoto S, Tsuchida N. Gene amplification and overexpression of epidermal growth factor receptor in squamous cell carcinoma of the head and neck. Head Neck. 1992;14(1):8–13. doi: 10.1002/hed.2880140103. [DOI] [PubMed] [Google Scholar]
- 5.Jarmuz-Szymczak M, Pelinska K, Kostrzewska-Poczekaj M, Bembnista E, Giefing M, Brauze D, et al. Heterogeneity of 11q13 region rearrangements in laryngeal squamous cell carcinoma analyzed by microarray platforms and fluorescence in situ hybridization. Mol Biol Rep. 2013;40(7):4161–71. doi: 10.1007/s11033-013-2496-4. [DOI] [PubMed] [Google Scholar]
- 6.Kostrzewska-Poczekaj M, Giefing M, Jarmuz M, Brauze D, Pelinska K, Grenman R, Bartochowska A, Szyfter W, and Szyfter K, “Recurrent amplification in the 22q11 region in laryngeal squamous cell carcinoma results in overexpression of the CRKL but not the MAPK1 oncogene,” Cancer Biomark. Sect. Dis. Markers, vol. 8, no. 1, pp. 11–19, 2011 2010. [DOI] [PubMed]
- 7.Nigg EA. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. BioEssays News Rev Mol Cell Dev Biol. 1995;17(6):471–80. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 8.Diril MK, Ratnacaram CK, Padmakumar VC, Du T, Wasser M, Coppola V, et al. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc Natl Acad Sci U S A. 2012;109(10):3826–31. doi: 10.1073/pnas.1115201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ford HL, Pardee AB. Cancer and the cell cycle. J Cell Biochem. 1999;75(S32):166–72. doi: 10.1002/(SICI)1097-4644(1999)75:32+<166::AID-JCB20>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 10.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11(3):961. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448(7155):811–5. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 12.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6(4):313–9. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 13.Meeker TC, Nagarajan L, Ar-Rushdi A, Rovera G, Huebner K, Croce CM. Characterization of the human PIM-1 gene: a putative proto-oncogene coding for a tissue specific member of the protein kinase family. Oncogene Res. 1987;1(1):87–101. [PubMed] [Google Scholar]
- 14.Malinen M, Jääskeläinen T, Pelkonen M, Heikkinen S, Väisänen S, Kosma VM, et al. Proto-oncogene PIM-1 is a novel estrogen receptor target associating with high grade breast tumors. Mol Cell Endocrinol. 2013;365(2):270–6. doi: 10.1016/j.mce.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 15.Enserink JM, Kolodner RD. An overview of Cdk1-controlled targets and processes. Cell Div. 2010;5:11. doi: 10.1186/1747-1028-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J-Q, Liu H-X, Liang Z, Sun Y-M, Wu M. Over-expression of p53, p21 and Cdc2 in histologically negative surgical margins is correlated with local recurrence of laryngeal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7(7):4295–302. [PMC free article] [PubMed] [Google Scholar]
- 17.Lian M, Fang J, Han D, Ma H, Feng L, Wang R, et al. Microarray gene expression analysis of tumorigenesis and regional lymph node metastasis in laryngeal squamous cell carcinoma. PLoS One. 2013;8(12):e84854. doi: 10.1371/journal.pone.0084854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, et al. Towards a knowledge-based human protein atlas. Nat Biotechnol. 2010;28(12):1248–50. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 19.Jarmuz M, Golusinski W, Grénman R, Szyfter K. Analysis of chromosome aberrations in cell lines derived from laryngeal cancer in relation to tumor progression. Eur Arch Oto-Rhino-Laryngol Off J Eur Fed Oto-Rhino-Laryngol Soc EUFOS Affil Ger Soc Oto-Rhino-Laryngol - Head Neck Surg. 2002;259(5):269–73. doi: 10.1007/s00405-002-0450-5. [DOI] [PubMed] [Google Scholar]
- 20.Jarmuz M, Grenman R, Golusinski W, Szyfter K. Aberrations of 11q13 in laryngeal squamous cell lines and their prognostic significance. Cancer Genet Cytogenet. 2005;160(1):82–8. doi: 10.1016/j.cancergencyto.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc. 2006;1(2):581–5. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- 22.Szaumkessel M, Richter J, Giefing M, Jarmuz M, Kiwerska K, Tönnies H, et al. Pyrosequencing-based DNA methylation profiling of Fanconi anemia/BRCA pathway genes in laryngeal squamous cell carcinoma. Int J Oncol. 2011;39(2):505–14. doi: 10.3892/ijo.2011.1039. [DOI] [PubMed] [Google Scholar]
- 23.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 24.Giefing M, Zemke N, Brauze D, Kostrzewska-Poczekaj M, Luczak M, Szaumkessel M, et al. High resolution ArrayCGH and expression profiling identifies PTPRD and PCDH17/PCH68 as tumor suppressor gene candidates in laryngeal squamous cell carcinoma. Genes Chromosomes Cancer. 2011;50(3):154–66. doi: 10.1002/gcc.20840. [DOI] [PubMed] [Google Scholar]
- 25.Giefing M, Martin-Subero JI, Kiwerska K, Jarmuz M, Grenman R, Siebert R, et al. Characterization of homozygous deletions in laryngeal squamous cell carcinoma cell lines. Cancer Genet Cytogenet. 2008;184(1):38–43. doi: 10.1016/j.cancergencyto.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Bodnar M, Szylberg Ł, Kaźmierczak W, Marszałek A. Immunohistochemical expression of p27(kip1) in metastatic laryngeal squamous cell carcinoma. Adv Med Sci. 2014;59(2):206–12. doi: 10.1016/j.advms.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 27.Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathol. 1987;8(3):138–40. [PubMed] [Google Scholar]
- 28.Bodnar M, Szylberg L, Kazmierczak W, Marszalek A. Differentiated expression of membrane type metalloproteinases (MMP-14, MMP-15) and pro-MMP2 in laryngeal squamous cell carcinoma. A novel mechanism. J Oral Pathol Med Off Publ Int Assoc Oral Pathol Am Acad Oral Pathol. 2013;42(3):267–74. doi: 10.1111/jop.12000. [DOI] [PubMed] [Google Scholar]
- 29.Bodnar M, Szylberg Ł, Kazmierczak W, Marszalek A. Tumor progression driven by pathways activating matrix metalloproteinases and their inhibitors. J Oral Pathol Med Off Publ Int Assoc Oral Pathol Am Acad Oral Pathol. 2015;44(6):437–43. doi: 10.1111/jop.12270. [DOI] [PubMed] [Google Scholar]
- 30.Burduk PK, Bodnar M, Sawicki P, Szylberg Ł, Wiśniewska E, Kaźmierczak W, et al. Expression of metalloproteinases 2 and 9 and tissue inhibitors 1 and 2 as predictors of lymph node metastases in oropharyngeal squamous cell carcinoma. Head Neck. 2015;37(3):418–22. doi: 10.1002/hed.23618. [DOI] [PubMed] [Google Scholar]
- 31.Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28(33):2925–39. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 32.Landry BD, Mapa CE, Arsenault HE, Poti KE, Benanti JA. Regulation of a transcription factor network by Cdk1 coordinates late cell cycle gene expression. EMBO J. 2014;33(9):1044–60. doi: 10.1002/embj.201386877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu P, Kao TP, Huang H. CDK1 promotes cell proliferation and survival via phosphorylation and inhibition of FOXO1 transcription factor. Oncogene. 2008;27(34):4733–44. doi: 10.1038/onc.2008.104. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes F, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348(11):994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 35.Blackwell KL, Burstein HJ, Storniolo AM, Rugo HS, Sledge G, Aktan G, et al. Overall survival benefit with lapatinib in combination with trastuzumab for patients with human epidermal growth factor receptor 2-positive metastatic breast cancer: final results from the EGF104900 study. J Clin Oncol Off J Am Soc Clin Oncol. 2012;30(21):2585–92. doi: 10.1200/JCO.2011.35.6725. [DOI] [PubMed] [Google Scholar]
- 36.Pezzuto F, Buonaguro L, Caponigro F, Ionna F, Starita N, Annunziata C, et al. Update on head and neck cancer: current knowledge on epidemiology, risk factors, molecular features and novel therapies. Oncology. 2015;89(3):125–36. doi: 10.1159/000381717. [DOI] [PubMed] [Google Scholar]
- 37.Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med. 2007;13(7):820–7. doi: 10.1038/nm1606. [DOI] [PubMed] [Google Scholar]
- 38.Linton A, Cheng YY, Griggs K, Kirschner MB, Gattani S, Srikaran S, et al. An RNAi-based screen reveals PLK1, CDK1 and NDC80 as potential therapeutic targets in malignant pleural mesothelioma. Br J Cancer. 2014;110(2):510–9. doi: 10.1038/bjc.2013.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xi Q, Huang M, Wang Y, Zhong J, Liu R, Xu G, et al. The expression of CDK1 is associated with proliferation and can be a prognostic factor in epithelial ovarian cancer. Tumor Biol. 2015;36(7):4939–48. doi: 10.1007/s13277-015-3141-8. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Zhu Y-H, Mao C-Q, Dou S, Shen S, Tan Z-B, et al. Triple negative breast cancer therapy with CDK1 siRNA delivered by cationic lipid assisted PEG-PLA nanoparticles. J Control Release Off J Control Release Soc. 2014;192:114–21. doi: 10.1016/j.jconrel.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Kang J, Sergio CM, Sutherland RL, Musgrove EA. Targeting cyclin-dependent kinase 1 (CDK1) but not CDK4/6 or CDK2 is selectively lethal to MYC-dependent human breast cancer cells. BMC Cancer. 2014;14:32. doi: 10.1186/1471-2407-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang JT, Wang H-M, Chang K-W, Chen W-H, Wen M-C, Hsu Y-M, et al. Identification of differentially expressed genes in oral squamous cell carcinoma (OSCC): overexpression of NPM, CDK1 and NDRG1 and underexpression of CHES1. Int J Cancer J Int Cancer. 2005;114(6):942–9. doi: 10.1002/ijc.20663. [DOI] [PubMed] [Google Scholar]
- 43.Chae SW, Sohn JH, Kim D-H, Choi YJ, Park YL, Kim K, et al. Overexpressions of Cyclin B1, cdc2, p16 and p53 in human breast cancer: the clinicopathologic correlations and prognostic implications. Yonsei Med J. 2011;52(3):445–53. doi: 10.3349/ymj.2011.52.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao J, Han S-X, Ma J-L, Ying X, Liu P, Li J, et al. The role of CDK1 in apoptin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2013;30(1):253–9. doi: 10.3892/or.2013.2426. [DOI] [PubMed] [Google Scholar]
- 45.Timofeev O, Cizmecioglu O, Settele F, Kempf T, Hoffmann I. Cdc25 phosphatases are required for timely assembly of CDK1-cyclin B at the G2/M transition. J Biol Chem. 2010;285(22):16978–90. doi: 10.1074/jbc.M109.096552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Oijen MGCT, Slootweg PJ. Oral field cancerization: carcinogen-induced independent events or micrometastatic deposits? Cancer Epidemiol Biomarkers Prev. 2000;9(3):249–56. [PubMed] [Google Scholar]
- 47.Chen X, Zhang F-H, Chen Q-E, Wang Y-Y, Wang Y-L, He J-C, et al. The clinical significance of CDK1 expression in oral squamous cell carcinoma. Med Oral Patol Oral Cir Bucal. 2015;20(1):e7–12. doi: 10.4317/medoral.19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sung W-W, Lin Y-M, Wu P-R, Yen H-H, Lai H-W, Su T-C, et al. High nuclear/cytoplasmic ratio of Cdk1 expression predicts poor prognosis in colorectal cancer patients. BMC Cancer. 2014;14(1):1–7. doi: 10.1186/1471-2407-14-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glover AR, Zhao JT, Gill AJ, Weiss J, Mugridge N, Kim E, Feeney AL, Ip JC, Reid G, Clarke S, Soon PSH, Robinson BG, Brahmbhatt H, MacDiarmid JA and Sidhu SB, “microRNA-7 as a tumor suppressor and novel therapeutic for adrenocortical carcinoma,” Oncotarget, Oct. 2015. [DOI] [PMC free article] [PubMed]
- 50.Chen S, Chen X, Xiu Y-L, Sun K-X, Zhao Y. MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial carcinoma tumorigenesis and progression. Cancer Lett. 2015;362(1):122–30. doi: 10.1016/j.canlet.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 51.Nair J, Jain P, Chandola U, Palve V, Vardhan NRH, Reddy RB, et al. Gene and miRNA expression changes in squamous cell carcinoma of larynx and hypopharynx. Genes Cancer. 2015;6(7–8):328–40. doi: 10.18632/genesandcancer.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karatas OF, Yuceturk B, Suer I, Yilmaz M, Cansiz H, Solak M, Ittmann M, and Ozen M, “Role of miR-145 in human laryngeal squamous cell carcinoma,” Head Neck, Oct. 2014. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 18 kb)
(DOCX 14 kb)
(DOCX 15 kb)
(DOCX 15 kb)